1WAE
 
 | | Crystal structure of H129V Mutant of Alcaligenes Xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Ellis, M.J, Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2004-10-26 | | Release date: | 2004-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Observation of an Unprecedented Cu Bis-His Site: Crystal Structure of the H129V Mutant of Nitrite Reductase
Inorg.Chem., 43, 2004
|
|
1GS7
 
 | | Crystal structure of H254F mutant of Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
1GS6
 
 | | Crystal structure of M144A mutant of Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, MAGNESIUM ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
1GS8
 
 | | Crystal structure of mutant D92N Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
1HAU
 
 | | X-RAY STRUCTURE OF A BLUE COPPER NITRITE REDUCTASE AT HIGH PH AND IN COPPER FREE FORM AT 1.9 A RESOLUTION | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE | | Authors: | Ellis, M.J, Dodd, F.E, Strange, R.W, Prudencio, M, Sawerseady, R.R, Hasnain, S.S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray Structure of a Blue Copper Nitrite Reductase at High Ph and in Copper-Free Form at 1.9 A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HAW
 
 | | X-RAY STRUCTURE OF A BLUE COPPER NITRITE REDUCTASE AT HIGH PH AND IN COPPER FREE FORM AT 1.9 A RESOLUTION | | Descriptor: | COPPER (I) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE | | Authors: | Ellis, M.J, Dodd, F.E, Strange, R.W, Prudencio, M, Sawerseady, R.R, Hasnain, S.S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray Structure of a Blue Copper Nitrite Reductase at High Ph and in Copper-Free Form at 1.9 A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1OE1
 
 | |
1OE3
 
 | | Atomic resolution structure of 'Half Apo' NiR | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL | | Authors: | Ellis, M.J, Dodd, F.E, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2003-03-18 | | Release date: | 2004-07-21 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic Resolution Structures of Native Copper Nitrite Reductase from Alcaligenes Xylosoxidans and the Active Site Mutant Asp92Glu
J.Mol.Biol., 328, 2003
|
|
1OE2
 
 | | Atomic Resolution Structure of D92E Mutant of Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL | | Authors: | Ellis, M.J, Dodd, F.E, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Atomic Resolution Structures of Native Copper Nitrite Reductase from Alcaligenes Xylosoxidans and the Active Site Mutant Asp92Glu
J.Mol.Biol., 328, 2003
|
|
2VW6
 
 | | NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS - 3 OF 3 | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL, ... | | Authors: | Ellis, M.J, Buffey, S.G, Hough, M.A, Hasnain, S.S. | | Deposit date: | 2008-06-16 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On-Line Optical and X-Ray Spectroscopies with Crystallography: An Integrated Approach for Determining Metalloprotein Structures in Functionally Well Defined States.
J.Synchrotron Radiat., 15, 2008
|
|
2VW7
 
 | | Nitrite reductase from Alcaligenes xylosoxidans - 1 of 3 | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL, ... | | Authors: | Ellis, M.J, Buffey, S.G, Hough, M.A, Hasnain, S.S. | | Deposit date: | 2008-06-16 | | Release date: | 2008-09-09 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On-Line Optical and X-Ray Spectroscopies with Crystallography: An Integrated Approach for Determining Metalloprotein Structures in Functionally Well Defined States.
J.Synchrotron Radiat., 15, 2008
|
|
2VW4
 
 | | NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS - 2 OF 3 | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL, ... | | Authors: | Ellis, M.J, Buffey, S.G, Hough, M.A, Hasnain, S.S. | | Deposit date: | 2008-06-16 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On-Line Optical and X-Ray Spectroscopies with Crystallography: An Integrated Approach for Determining Metalloprotein Structures in Functionally Well Defined States.
J.Synchrotron Radiat., 15, 2008
|
|
6BDG
 
 | | HFQ monomer in spacegroup p6 at 1.93 angstrom resolution | | Descriptor: | RNA-binding protein Hfq | | Authors: | Brown, C, Zhang, K, Seo, C, Ellis, M.J, Hanniford, D.B, Junop, M. | | Deposit date: | 2017-10-23 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | HFQ monomer in spacegroup p6 at 1.93 angstrom resolution
To Be Published
|
|
3KBB
 
 | | Crystal structure of putative beta-phosphoglucomutase from Thermotoga maritima | | Descriptor: | GLYCEROL, Phosphorylated carbohydrates phosphatase TM_1254, SULFATE ION | | Authors: | Strange, R.W, Antonyuk, S.V, Ellis, M.J, Bessho, Y, Kuramitsu, S, Yokoyama, S, Hasnain, S.S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of a putative beta-phosphoglucomutase (TM1254) from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3IXQ
 
 | | Structure of ribose 5-phosphate isomerase a from methanocaldococcus jannaschii | | Descriptor: | ACETATE ION, CHLORIDE ION, Ribose-5-phosphate isomerase A, ... | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-04 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of an archaeal ribose-5-phosphate isomerase from Methanocaldococcus jannaschii (MJ1603).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3IWT
 
 | | Structure of hypothetical molybdenum cofactor biosynthesis protein B from Sulfolobus tokodaii | | Descriptor: | 178aa long hypothetical molybdenum cofactor biosynthesis protein B, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-03 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of hypothetical Mo-cofactor biosynthesis protein B (ST2315) from Sulfolobus tokodaii
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3KB6
 
 | | Crystal structure of D-Lactate dehydrogenase from aquifex aeolicus complexed with NAD and Lactic acid | | Descriptor: | D-lactate dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Ellis, M.J, Bessho, Y, Kuramitsu, S, Yokoyama, S, Hasnain, S.S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of D-lactate dehydrogenase from Aquifex aeolicus complexed with NAD(+) and lactic acid (or pyruvate).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2BO0
 
 | | Crystal structure of the C130A mutant of nitrite reductase from Alcaligenes xylosoxidans | | Descriptor: | DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Hough, M.A, Ellis, M.J, Antonyuk, S, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-04-06 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High Resolution Structural Studies of Mutants Provide Insights Into Catalysis and Electron Transfer Processes in Copper Nitrite Reductase
J.Mol.Biol., 350, 2005
|
|
2BP8
 
 | | M144Q Structure of nitrite reductase from Alcaligenes xylosoxidans | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, SULFATE ION, ... | | Authors: | Hough, M.A, Ellis, M.J, Antonyuk, S, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-04-18 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Resolution Structural Studies of Mutants Provide Insights Into Catalysis and Electron Transfer Processes in Copper Nitrite Reductase
J.Mol.Biol., 350, 2005
|
|
2BP0
 
 | | M144L mutant of nitrite reductase from Alcaligenes xylosoxidans | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, SULFATE ION, ... | | Authors: | Hough, M.A, Ellis, M.J, Antonyuk, S, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-04-17 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Resolution Structural Studies of Mutants Provide Insights Into Catalysis and Electron Transfer Processes in Copper Nitrite Reductase
J.Mol.Biol., 350, 2005
|
|
2CAL
 
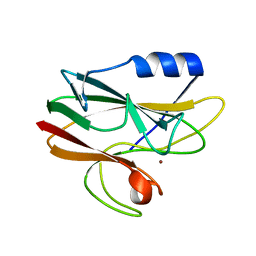 | | Crystal structure of His143Met rusticyanin | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Barrett, M.L, Harvey, I, Sundararajan, M, Surendran, R, Hall, J.F, Ellis, M.J, Hough, M.A, Strange, R.W, Hillier, I.H, Hasnain, S.S. | | Deposit date: | 2005-12-21 | | Release date: | 2006-01-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Crystal Structures, Exafs, and Quantum Chemical Studies of Rusticyanin and its Two Mutants Provide Insight Into its Unusual Properties.
Biochemistry, 45, 2006
|
|
2CAK
 
 | | 1.27Angstrom Structure of Rusticyanin from Thiobacillus ferrooxidans | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Barrett, M.L, Harvey, I, Sundararajan, M, Surendran, R, Hall, J.F, Ellis, M.J, Hough, M.A, Strange, R.W, Hillier, I.H, Hasnain, S.S. | | Deposit date: | 2005-12-21 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Atomic Resolution Crystal Structures, Exafs, and Quantum Chemical Studies of Rusticyanin and its Two Mutants Provide Insight Into its Unusual Properties.
Biochemistry, 45, 2006
|
|
2PCR
 
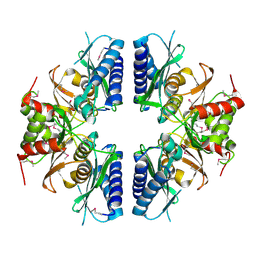 | | Crystal structure of Myo-inositol-1(or 4)-monophosphatase (aq_1983) from Aquifex Aeolicus VF5 | | Descriptor: | Inositol-1-monophosphatase | | Authors: | Jeyakanthan, J, Gayathri, D, Velmurugan, D, Agari, Y, Bessho, Y, Ellis, M.J, Antonyuk, S.V, Strange, R.W, Hasnain, S.S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Myo-inositol-1(or 4)-monophosphatase (aq_1983) from Aquifex Aeolicus VF5
To be Published
|
|
2WQK
 
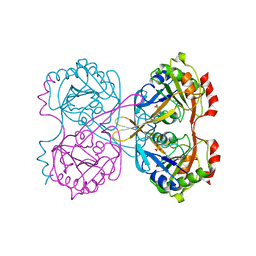 | | Crystal Structure of Sure Protein from Aquifex aeolicus | | Descriptor: | 5'-NUCLEOTIDASE SURE, SODIUM ION, SULFATE ION | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-08-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Sure Protein from Aquifex Aeolicus Vf5 at 1.5 A Resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2YV3
 
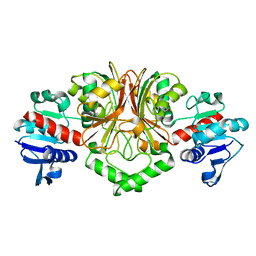 | | Crystal Structure of Aspartate Semialdehyde Dehydrogenase from Thermus thermophilus HB8 | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Kagawa, W, Fujikawa, N, Kurumizaka, H, Bessho, Y, Ellis, M.J, Antonyuk, S.V, Strange, R.W, Hasnain, S.S, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-07 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Aspartate Semialdehyde Dehydrogenase from Thermus thermophilus HB8
To be Published
|
|
