1AE9
 
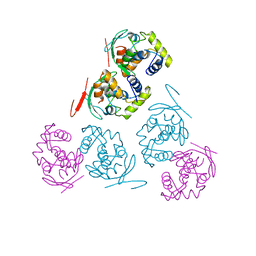 | |
1BNK
 
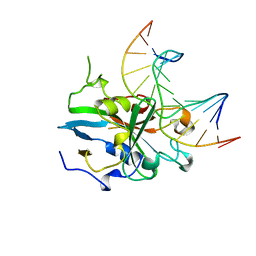 | | HUMAN 3-METHYLADENINE DNA GLYCOSYLASE COMPLEXED TO DNA | | Descriptor: | DNA (5'-D(*GP*AP*CP*AP*TP*GP*YRRP*TP*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), PROTEIN (3-METHYLADENINE DNA GLYCOSYLASE) | | Authors: | Lau, A.Y, Schaerer, O.D, Samson, L, Verdine, G.L, Ellenberger, T. | | Deposit date: | 1998-07-29 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision.
Cell(Cambridge,Mass.), 95, 1998
|
|
1CR4
 
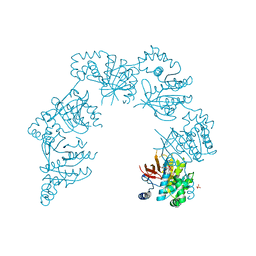 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTDP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CR1
 
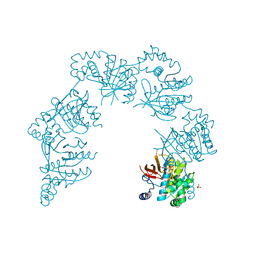 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTTP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CR2
 
 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA PRIMASE/HELICASE, SULFATE ION | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1D5Y
 
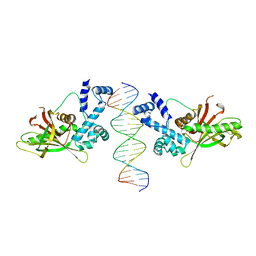 | | CRYSTAL STRUCTURE OF THE E. COLI ROB TRANSCRIPTION FACTOR IN COMPLEX WITH DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*CP*AP*TP*TP*CP*AP*GP*TP*GP*CP*TP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*CP*AP*GP*CP*AP*CP*TP*GP*AP*AP*TP*GP*TP*CP*AP*AP*AP*G)-3'), ROB TRANSCRIPTION FACTOR | | Authors: | Kwon, H.J, Bennik, M.H.J, Demple, B, Ellenberger, T. | | Deposit date: | 1999-10-12 | | Release date: | 2000-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA.
Nat.Struct.Biol., 7, 2000
|
|
1CR0
 
 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE4 PROTEIN OF BACTERIOPHAGE T7 | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
6WH1
 
 | | Structure of the complex of human DNA ligase III-alpha and XRCC1 BRCT domains | | Descriptor: | DNA ligase 3 alpha, X-ray repair cross complementing protein 1 variant | | Authors: | Pourfarjam, Y, Ellenberger, T, Tainer, J.A, Tomkinson, A.E, Kim, I.K. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase III alpha within a flexible DNA repair complex.
Nucleic Acids Res., 49, 2021
|
|
6WH2
 
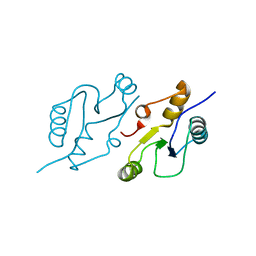 | | Structure of the C-terminal BRCT domain of human XRCC1 | | Descriptor: | X-ray repair cross complementing protein 1 variant | | Authors: | Pourfarjam, Y, Ellenberger, T, Tainer, J.A, Tomkinson, A.E, Kim, I.K. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase III alpha within a flexible DNA repair complex.
Nucleic Acids Res., 49, 2021
|
|
3PNT
 
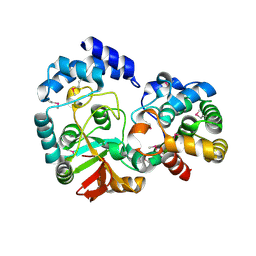 | | Crystal Structure of the Streptococcus pyogenes NAD+ glycohydrolase SPN in complex with IFS, the Immunity Factor for SPN | | Descriptor: | Immunity factor for SPN, NAD+-glycohydrolase | | Authors: | Smith, C.L, Stine Elam, J, Ellenberger, T, Ghosh, J, Pinkner, J.S, Hultgren, S.J, Caparon, M.G. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Streptococcus pyogenes Immunity to Its NAD(+) Glycohydrolase Toxin.
Structure, 19, 2011
|
|
3QB2
 
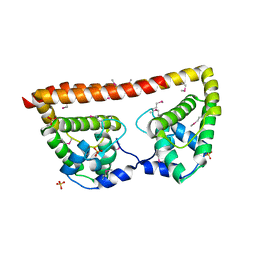 | |
3L2P
 
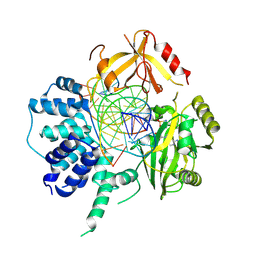 | | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching Between Two DNA Bound States | | Descriptor: | 5'-D(*GP*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*CP*CP*G)-3', 5'-D(*GP*TP*CP*GP*GP*AP*CP*TP*G)-3', 5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*GP*TP*C)-3', ... | | Authors: | Cotner-Gohara, E.A, Kim, I.K, Hammel, M, Tainer, J.A, Tomkinson, A, Ellenberger, T. | | Deposit date: | 2009-12-15 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching between Two DNA-Bound States.
Biochemistry, 49, 2010
|
|
5IKN
 
 | |
5KVH
 
 | | Crystal structure of human apoptosis-inducing factor with W196A mutation | | Descriptor: | Apoptosis-inducing factor 1, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brosey, C.A, Nix, J, Ellenberger, T, Tainer, J.A. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Defining NADH-Driven Allostery Regulating Apoptosis-Inducing Factor.
Structure, 24, 2016
|
|
5KVI
 
 | | Crystal structure of monomeric human apoptosis-inducing factor with E413A/R422A/R430A mutations | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Apoptosis-inducing factor 1, mitochondrial, ... | | Authors: | Brosey, C.A, Nix, J, Ellenberger, T, Tainer, J.A. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Defining NADH-Driven Allostery Regulating Apoptosis-Inducing Factor.
Structure, 24, 2016
|
|
2ONT
 
 | | A swapped dimer of the HIV-1 capsid C-terminal domain | | Descriptor: | Capsid protein p24 | | Authors: | Ivanov, D, Tsodikov, O.V, Kasanov, J, Ellenberger, T, Wagner, G, Collins, T. | | Deposit date: | 2007-01-24 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Domain-swapped dimerization of the HIV-1 capsid C-terminal domain
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3UEK
 
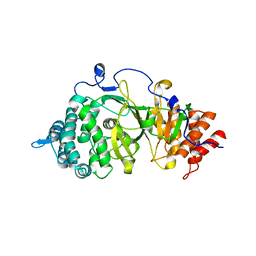 | | Crystal structure of the catalytic domain of rat poly (ADP-ribose) glycohydrolase | | Descriptor: | Poly(ADP-ribose) glycohydrolase | | Authors: | Kim, I.K, Kiefer, J.R, Stegemann, R.A, Classen, S, Tainer, J.A, Ellenberger, T. | | Deposit date: | 2011-10-30 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UEL
 
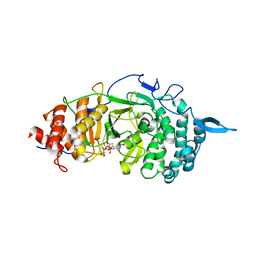 | | Crystal structure of the catalytic domain of rat poly (ADP-ribose) glycohydrolase bound to ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Poly(ADP-ribose) glycohydrolase | | Authors: | Kim, I.K, Kiefer, J.R, Stegemann, R.A, Classen, S, Tainer, J.A, Ellenberger, T. | | Deposit date: | 2011-10-30 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1E0K
 
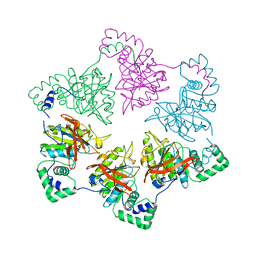 | | gp4d helicase from phage T7 | | Descriptor: | DNA HELICASE | | Authors: | Singleton, M.R, Sawaya, M.R, Ellenberger, T, Wigley, D.B. | | Deposit date: | 2000-03-30 | | Release date: | 2000-06-09 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of T7 Gene 4 Ring Helicase Indicates a Mechanism for Sequential Hydrolysis of Nucleotides
Cell(Cambridge,Mass.), 101, 2000
|
|
1E0J
 
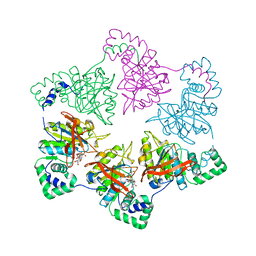 | | gp4d helicase from phage T7 ADPNP complex | | Descriptor: | DNA HELICASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Singleton, M.R, Sawaya, M.R, Ellenberger, T, Wigley, D.B. | | Deposit date: | 2000-03-30 | | Release date: | 2000-06-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of T7 Gene 4 Ring Helicase Indicates a Mechanism for Sequential Hydrolysis of Nucleotides
Cell(Cambridge,Mass.), 101, 2000
|
|
1EWN
 
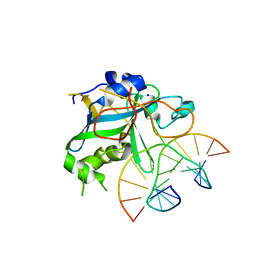 | | CRYSTAL STRUCTURE OF THE HUMAN AAG DNA REPAIR GLYCOSYLASE COMPLEXED WITH 1,N6-ETHENOADENINE-DNA | | Descriptor: | 3-METHYL-ADENINE DNA GLYCOSYLASE, DNA (5'-D(*GP*AP*CP*AP*TP*GP*(EDA)P*TP*TP*GP*CP*C)-3'), DNA (5'-D(P*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), ... | | Authors: | Lau, A.Y, Wyatt, M.D, Glassner, B.J, Samson, L.D, Ellenberger, T. | | Deposit date: | 2000-04-26 | | Release date: | 2000-12-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F4R
 
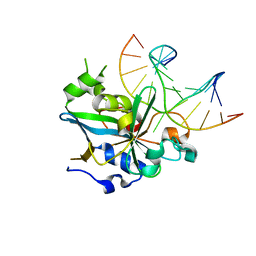 | | CRYSTAL STRUCTURE OF THE HUMAN AAG DNA REPAIR GLYCOSYLASE COMPLEXED WITH 1,N6-ETHENOADENINE-DNA | | Descriptor: | 3-METHYL-ADENINE DNA GLYCOSYLASE, DNA (5'-D(*GP*AP*CP*AP*TP*GP*(EDA)P*TP*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), ... | | Authors: | Lau, A.Y, Wyatt, M.D, Glassner, B.J, Samson, L.D, Ellenberger, T. | | Deposit date: | 2000-06-08 | | Release date: | 2000-12-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F6O
 
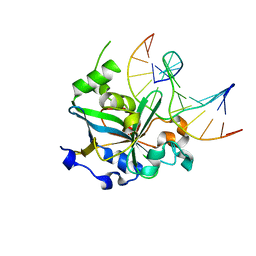 | | CRYSTAL STRUCTURE OF THE HUMAN AAG DNA REPAIR GLYCOSYLASE COMPLEXED WITH DNA | | Descriptor: | 3-METHYL-ADENINE DNA GLYCOSYLASE, DNA (5'-D(*GP*AP*CP*AP*TP*GP*(YRR)P*TP*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), ... | | Authors: | Lau, A.Y, Wyatt, M.D, Glassner, B.J, Samson, L.D, Ellenberger, T. | | Deposit date: | 2000-06-22 | | Release date: | 2000-12-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6OA3
 
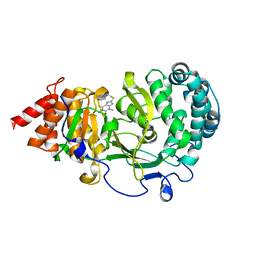 | | Structure of human PARG complexed with JA2131 | | Descriptor: | (8S)-1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-6-sulfanylidene-1,3,6,7,8,9-hexahydro-2H-purin-2-one, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
6O9Y
 
 | | Structure of human PARG complexed with JA2-8 | | Descriptor: | 7-[(2S)-2-hydroxy-3-(morpholin-4-yl)propyl]-1,3-dimethyl-3,7-dihydro-1H-purine-2,6-dione, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
