2B1F
 
 | | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat | | Descriptor: | General control protein GCN4 | | Authors: | Deng, Y, Liu, J, Zheng, Q, Eliezer, D, Kallenbach, N.R, Lu, M. | | Deposit date: | 2005-09-15 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat.
Structure, 14, 2006
|
|
2B22
 
 | | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat | | Descriptor: | General control protein GCN4, SODIUM ION | | Authors: | Deng, Y, Liu, J, Zheng, Q, Eliezer, D, Kallenbach, N.R, Lu, M. | | Deposit date: | 2005-09-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat.
Structure, 14, 2006
|
|
2FUN
 
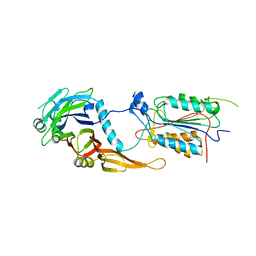 | | alternative p35-caspase-8 complex | | Descriptor: | Early 35 kDa protein, caspase-8 | | Authors: | Lu, M, Min, T, Eliezer, D, Wu, H. | | Deposit date: | 2006-01-27 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Native chemical ligation in covalent caspase inhibition by p35.
Chem.Biol., 13, 2006
|
|
5BRL
 
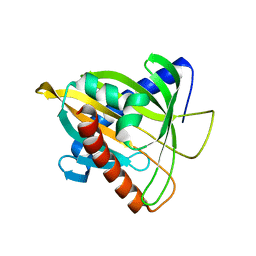 | | Crystal Structure of L124D STARD4 | | Descriptor: | StAR-related lipid transfer protein 4 | | Authors: | Iaea, D.B, Dikiy, I, Kiburu, I, Eliezer, D.E, Maxfield, F.R. | | Deposit date: | 2015-05-31 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | STARD4 Membrane Interactions and Sterol Binding.
Biochemistry, 54, 2015
|
|
5E1A
 
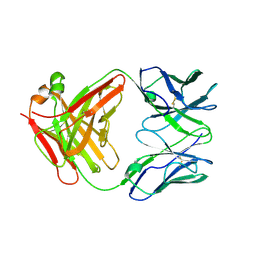 | | Structure of KcsA with L24C/R117C mutations | | Descriptor: | POTASSIUM ION, antibody Fab fragment heavy chain, antibody Fab fragment light chain, ... | | Authors: | Upadhyay, V, Kim, D.M, Nimigean, C.M. | | Deposit date: | 2015-09-29 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Conformational heterogeneity in closed and open states of the KcsA potassium channel in lipid bicelles.
J.Gen.Physiol., 148, 2016
|
|
6UWF
 
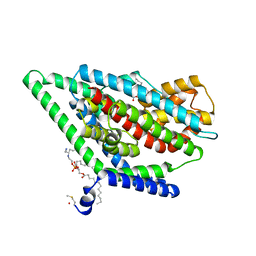 | | GltPh in complex with L-aspartate and sodium ions in outward-facing state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Wang, X, Boudker, O. | | Deposit date: | 2019-11-05 | | Release date: | 2020-06-03 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Use of paramagnetic 19 F NMR to monitor domain movement in a glutamate transporter homolog.
Nat.Chem.Biol., 16, 2020
|
|
3FX0
 
 | |
6UWL
 
 | |
6BYM
 
 | | Crystal structure of the sterol-bound second StART domain of yeast Lam4 | | Descriptor: | 25-HYDROXYCHOLESTEROL, Sterol-binding protein | | Authors: | Jentsch, J.A, Kiburu, I.N, Wu, J, Pandey, K, Boudker, O, Menon, A.K. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of sterol binding and transport by a yeast StARkin domain.
J. Biol. Chem., 293, 2018
|
|
6BYD
 
 | | Crystal structure of the second StART domain of yeast Lam4 | | Descriptor: | Membrane-anchored lipid-binding protein LAM4 | | Authors: | Jentsch, J.A, Kiburu, I.N, Wu, J, Pandey, K, Boudker, O, Menon, A.K. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Structural basis of sterol binding and transport by a yeast StARkin domain.
J. Biol. Chem., 293, 2018
|
|
7UGX
 
 | | Asp-bound GltPh RSMR mutant in IFS-B1 state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19 F NMR and Cryo-EM.
J.Am.Chem.Soc., 145, 2023
|
|
7UG0
 
 | | TBOA-bound GltPh RSMR mutant in IFS state | | Descriptor: | (3S)-3-(BENZYLOXY)-L-ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
7UGD
 
 | | Asp-bound GltPh RSMR mutant in IFS-A1 state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
7UH3
 
 | | Asp-bound GltPh RSMR mutant in iOFS state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
7UGJ
 
 | | TBOA-bound GltPh RSMR mutant in OFS state | | Descriptor: | (3S)-3-(BENZYLOXY)-L-ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
7UGV
 
 | | Asp-bound GltPh RSMR mutant in IFS-A2 state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19 F NMR and Cryo-EM.
J.Am.Chem.Soc., 145, 2023
|
|
7UH6
 
 | | Asp-bound GltPh RSMR mutant in IFS-B2 state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
