1BE4
 
 | | NUCLEOSIDE DIPHOSPHATE KINASE ISOFORM B FROM BOVINE RETINA | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, NUCLEOSIDE DIPHOSPHATE TRANSFERASE | | Authors: | Ladner, J.E, Abdulaev, N.G, Kakuev, D.L, Karaschuk, G.N, Tordova, M, Eisenstein, E, Fujiwara, J.H, Ridge, K.D, Gilliland, G.L. | | Deposit date: | 1998-05-19 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nucleoside diphosphate kinase from bovine retina: purification, subcellular localization, molecular cloning, and three-dimensional structure.
Biochemistry, 37, 1998
|
|
1TDJ
 
 | | THREONINE DEAMINASE (BIOSYNTHETIC) FROM E. COLI | | Descriptor: | BIOSYNTHETIC THREONINE DEAMINASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gallagher, D.T, Gilliland, G.L, Xiao, G, Eisenstein, E. | | Deposit date: | 1998-03-27 | | Release date: | 1998-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase.
Structure, 6, 1998
|
|
1XAX
 
 | | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae | | Descriptor: | Hypothetical UPF0054 protein HI0004 | | Authors: | Yeh, D.C, Parsons, J.F, Parsons, L.M, Liu, F, Eisenstein, E, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-26 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae, and comparison with the X-ray structure of an Aquifex aeolicus homolog
Protein Sci., 14, 2005
|
|
1MXI
 
 | | Structure of YibK from Haemophilus influenzae (HI0766): a Methyltransferase with a Cofactor Bound at a Site Formed by a Knot | | Descriptor: | Hypothetical tRNA/rRNA methyltransferase HI0766, IODIDE ION, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lim, K, Zhang, H, Tempczyk, A, Bonander, N, Toedt, J, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-10-02 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): a Cofactor Bound at a Site Formed by a Knot
Proteins, 51, 2003
|
|
1NF9
 
 | | Crystal Structure of PhzD protein from Pseudomonas aeruginosa | | Descriptor: | FORMIC ACID, octyl beta-D-glucopyranoside, phenazine biosynthesis protein phzD | | Authors: | Parsons, F, Calabrese, K, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2002-12-13 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and mechanism of Pseudomonas aeruginosa PhzD, an isochorismatase from the phenazine biosynthetic pathway
Biochemistry, 42, 2003
|
|
1NF8
 
 | | Crystal structure of PhzD protein active site mutant with substrate | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, octyl beta-D-glucopyranoside, phenazine biosynthesis protein phzD | | Authors: | Parsons, F, Calabrese, K, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2002-12-13 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of Pseudomonas aeruginosa PhzD, an isochorismatase from the phenazine biosynthetic pathway
Biochemistry, 42, 2003
|
|
1T9M
 
 | | X-ray crystal structure of phzG from pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Parsons, J.F, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2004-05-18 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the phenazine biosynthesis enzyme PhzG.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1T6K
 
 | | Crystal structure of phzF from Pseudomonas fluorescens 2-79 | | Descriptor: | Phenazine biosynthesis protein phzF, SULFATE ION | | Authors: | Parsons, J.F, Song, F, Parsons, L, Calabrese, K, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2004-05-06 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the phenazine biosynthesis protein PhzF from Pseudomonas fluorescens 2-79
Biochemistry, 43, 2004
|
|
1TY9
 
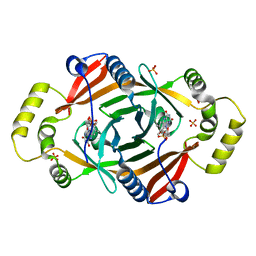 | | X-RAY CRYSTAL STRUCTURE OF PHZG FROM PSEUDOMONAS FLUORESCENS | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phenazine biosynthesis protein phzG, SULFATE ION | | Authors: | Parsons, J.F, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2004-07-07 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the phenazine biosynthesis enzyme PhzG.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1I2K
 
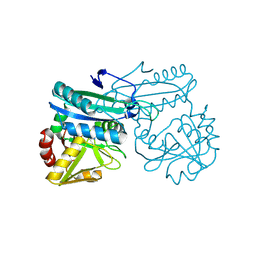 | | AMINODEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jensen, P.Y, Parsons, J.F, Fisher, K.E, Pachikara, A.S, Tordova, M, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-02-09 | | Release date: | 2003-09-02 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and Mechanism of Escherichia coli Aminodeoxychorismate Lyase
To be Published
|
|
1I2L
 
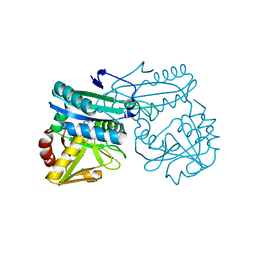 | | DEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI WITH INHIBITOR | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Jensen, P.Y, Parsons, J.F, Fisher, K.E, Pachikara, A.S, Tordova, M, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-02-09 | | Release date: | 2003-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Escherichia coli Aminodeoxychorismate Lyase
To be Published
|
|
1IM8
 
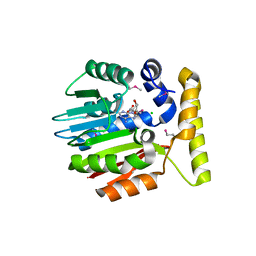 | | Crystal structure of YecO from Haemophilus influenzae (HI0319), a methyltransferase with a bound S-adenosylhomocysteine | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOSELENOCYSTEINE, YecO | | Authors: | Lim, K, Zhang, H, Tempczyk, A, Bonander, N, Toedt, J, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-10 | | Release date: | 2001-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of YecO from Haemophilus influenzae (HI0319) reveals a methyltransferase fold and a bound S-adenosylhomocysteine.
Proteins, 45, 2001
|
|
1J8B
 
 | | Structure of YbaB from Haemophilus influenzae (HI0442), a protein of unknown function | | Descriptor: | YbaB | | Authors: | Lim, K, Tempcyzk, A, Toedt, J, Parsons, J.F, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-21 | | Release date: | 2003-01-14 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of YbaB from Haemophilus influenzae (HI0442), a
protein of unknown function coexpressed with the recombinational
DNA repair protein RecR
Proteins, 50, 2003
|
|
1IMU
 
 | | Solution Structure of HI0257, a Ribosome Binding Protein | | Descriptor: | HYPOTHETICAL PROTEIN HI0257 | | Authors: | Parsons, L, Eisenstein, E, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-11 | | Release date: | 2001-10-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HI0257, a bacterial ribosome binding protein.
Biochemistry, 40, 2001
|
|
1J7H
 
 | | Solution Structure of HI0719, a Hypothetical Protein From Haemophilus Influenzae | | Descriptor: | HYPOTHETICAL PROTEIN HI0719 | | Authors: | Parsons, L, Bonander, N, Eisenstein, E, Gilson, M, Kairys, V, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-16 | | Release date: | 2003-02-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Functional Ligand Screening of HI0719, a Highly Conserved Protein from Bacteria to Humans in the YjgF/YER057c/UK114 Family
Biochemistry, 42, 2003
|
|
1J85
 
 | | Structure of YibK from Haemophilus influenzae (HI0766), a truncated sequence homolog of tRNA (guanosine-2'-O-) methyltransferase (SpoU) | | Descriptor: | YibK | | Authors: | Lim, K, Zhang, H, Toedt, J, Tempcyzk, A, Krajewski, W, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-20 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae
(HI0766): A cofactor bound at a site formed by a knot
Proteins, 51, 2003
|
|
1J9A
 
 | | OLIGORIBONUCLEASE | | Descriptor: | OLIGORIBONUCLEASE, SULFATE ION | | Authors: | Bonander, N, Tordova, M, Ladner, J.E, Eisenstein, E, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-24 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of Haemophilus Influenzae HI1715, an Oligoribonuclease
To be Published
|
|
1JN1
 
 | | Structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Haemophilus influenzae (HI0671) | | Descriptor: | 2C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE SYNTHASE, COBALT (II) ION, SULFATE ION | | Authors: | Lehmann, C, Lim, K, Toedt, J, Krajewski, W, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-21 | | Release date: | 2002-08-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of 2C-methyl-D-erythrol-2,4-cyclodiphosphate synthase from Haemophilus influenzae: activation by conformational transition.
Proteins, 49, 2002
|
|
1JJV
 
 | | DEPHOSPHO-COA KINASE IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DEPHOSPHO-COA KINASE, MERCURY (II) ION, ... | | Authors: | Obmolova, G, Teplyakov, A, Bonander, N, Eisenstein, E, Howard, A.J, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-09 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dephospho-coenzyme A kinase from Haemophilus influenzae.
J.Struct.Biol., 136, 2001
|
|
1JOE
 
 | | Crystal Structure of Autoinducer-2 Production Protein (LuxS) from Heamophilus influenzae | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN, MERCURY (II) ION, ZINC ION | | Authors: | Chen, C.C.H, Parsons, J.F, Lim, K, Lehmann, C, Tempczyk, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-27 | | Release date: | 2001-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CRYSTAL STRUCTURE OF AUTOINDUCER-2 PRODUCTION PROTEIN (LUXS) FROM HEAMOPHILUS INFLUENZAE--A CASE OF TWINNED CRYSTAL
To be Published
|
|
1K0G
 
 | | THE CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE SYNTHASE FROM PHOSPHATE GROWN CRYSTALS | | Descriptor: | PHOSPHATE ION, TRYPTOPHAN, p-aminobenzoate synthase component I | | Authors: | Parsons, J.F, Jensen, P.Y, Pachikara, A.S, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Escherichia coli aminodeoxychorismate synthase: architectural conservation and diversity in chorismate-utilizing enzymes.
Biochemistry, 41, 2002
|
|
1JOV
 
 | | Crystal Structure Analysis of HI1317 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HI1317, SULFATE ION | | Authors: | Bonander, N, Tordova, M, Howard, A.J, Eisenstein, E, Gilliland, G, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-31 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal 1.57-A Crystal Structure of HI1317
TO BE PUBLISHED
|
|
1JOS
 
 | | Ribosome Binding Factor A(rbfA) | | Descriptor: | RIBOSOME-BINDING FACTOR A | | Authors: | Bonander, N, Tordova, M, Howard, A.J, Eisenstein, E, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-30 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7-A Crystal Structure of HI1288 - Ribosome Binding Factor A (rbfA), a Cold Response Protein
To be Published
|
|
1K0E
 
 | | THE CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE SYNTHASE FROM FORMATE GROWN CRYSTALS | | Descriptor: | FORMIC ACID, TRYPTOPHAN, p-aminobenzoate synthase component I | | Authors: | Parsons, J.F, Jensen, P.Y, Pachikara, A.S, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli aminodeoxychorismate synthase: architectural conservation and diversity in chorismate-utilizing enzymes.
Biochemistry, 41, 2002
|
|
4QT8
 
 | | Crystal Structure of RON Sema-PSI-IPT1 extracellular domains in complex with MSP beta-chain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hepatocyte growth factor-like protein, Macrophage-stimulating protein receptor, ... | | Authors: | Herzberg, O, Chao, K.L. | | Deposit date: | 2014-07-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the binding specificity of human Recepteur d'Origine Nantais (RON) receptor tyrosine kinase to macrophage-stimulating protein.
J.Biol.Chem., 289, 2014
|
|
