4WRI
 
 | | Crystal structure of okadaic acid binding protein 2.1 | | Descriptor: | OKADAIC ACID, Okadaic acid binding protein 2-alpha | | Authors: | Ehara, H, Makino, M, Kodama, K, Ito, T, Sekine, S, Fukuzawa, S, Yokoyama, S, Tachibana, K. | | Deposit date: | 2014-10-24 | | Release date: | 2015-05-27 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Okadaic Acid Binding Protein 2.1: A Sponge Protein Implicated in Cytotoxin Accumulation
Chembiochem, 16, 2015
|
|
3AYH
 
 | |
5X51
 
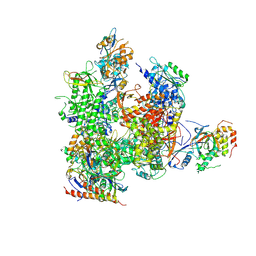 | | RNA Polymerase II from Komagataella Pastoris (Type-3 crystal) | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, RNA polymerase II subunit, ... | | Authors: | Ehara, H, Umehara, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-17 | | Method: | X-RAY DIFFRACTION (6.996 Å) | | Cite: | Crystal structure of RNA polymerase II from Komagataella pastoris
Biochem. Biophys. Res. Commun., 487, 2017
|
|
5X50
 
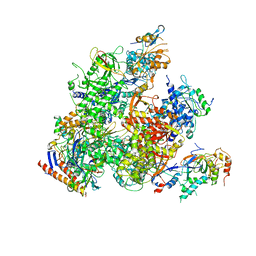 | | RNA Polymerase II from Komagataella Pastoris (Type-2 crystal) | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, RNA polymerase II subunit, ... | | Authors: | Ehara, H, Umehara, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.293 Å) | | Cite: | Crystal structure of RNA polymerase II from Komagataella pastoris
Biochem. Biophys. Res. Commun., 487, 2017
|
|
5XON
 
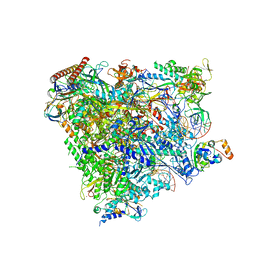 | | RNA Polymerase II elongation complex bound with Spt4/5 and TFIIS | | Descriptor: | DNA (48-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Yokoyama, T, Shigematsu, H, Shirouzu, M, Sekine, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structure of the complete elongation complex of RNA polymerase II with basal factors
Science, 357, 2017
|
|
6IR9
 
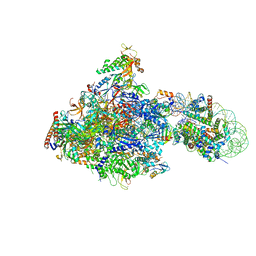 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6J50
 
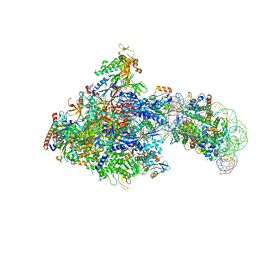 | | RNA polymerase II elongation complex bound with Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome (tilted conformation) | | Descriptor: | DNA (198-MER), DNA (41-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6J4W
 
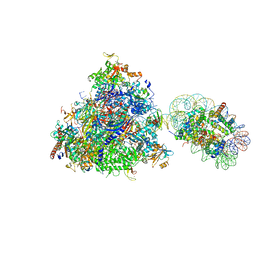 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6J4X
 
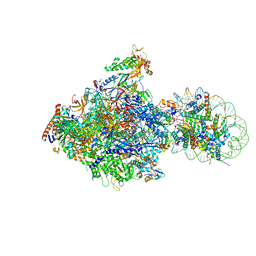 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-1) of the nucleosome (+1A) | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6J4Z
 
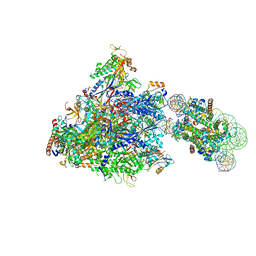 | | RNA polymerase II elongation complex bound with Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA (42-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6J4Y
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-1) of the nucleosome (+1B) | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6J51
 
 | | RNA polymerase II elongation complex bound with Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome, weak Elf1 (+1 position) | | Descriptor: | DNA (198-MER), DNA (36-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
5XOG
 
 | | RNA Polymerase II elongation complex bound with Spt5 KOW5 and Elf1 | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA (30-MER), DNA (39-MER), ... | | Authors: | Ehara, H, Shirouzu, M, Sekine, S. | | Deposit date: | 2017-05-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the complete elongation complex of RNA polymerase II with basal factors
Science, 357, 2017
|
|
5X4Z
 
 | | RNA Polymerase II from Komagataella Pastoris (Type-1 crystal) | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, RNA polymerase II subunit, ... | | Authors: | Ehara, H, Umehara, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-17 | | Method: | X-RAY DIFFRACTION (7.8 Å) | | Cite: | Crystal structure of RNA polymerase II from Komagataella pastoris
Biochem. Biophys. Res. Commun., 487, 2017
|
|
7XSZ
 
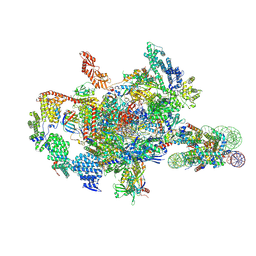 | | RNA polymerase II elongation complex transcribing a nucleosome (EC115) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-15 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XT7
 
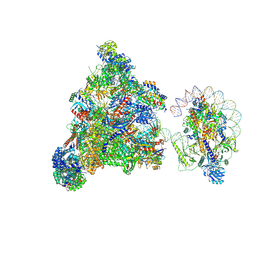 | | RNA polymerase II elongation complex transcribing a nucleosome (EC49B) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-16 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XTI
 
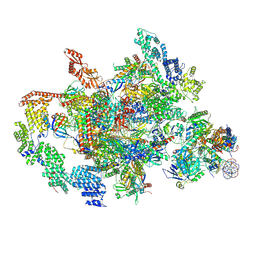 | | RNA polymerase II elongation complex transcribing a nucleosome (EC58hex) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XTD
 
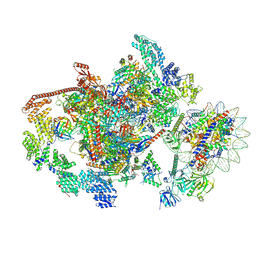 | | RNA polymerase II elongation complex transcribing a nucleosome (EC58oct) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-16 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XN7
 
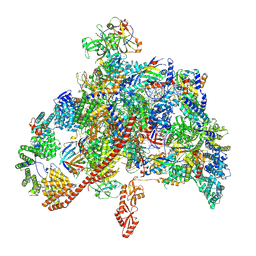 | | RNA polymerase II elongation complex containing Spt4/5, Elf1, Spt6, Spn1 and Paf1C | | Descriptor: | Chromatin elongation factor SPT5, Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XSX
 
 | | RNA polymerase II elongation complex transcribing a nucleosome (EC49) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-15 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XSE
 
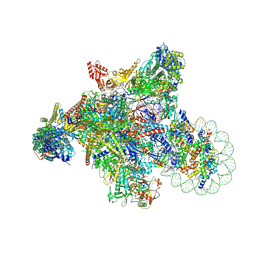 | | RNA polymerase II elongation complex transcribing a nucleosome (EC42) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
5B0U
 
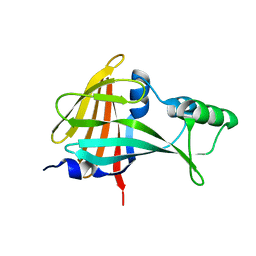 | |
8HE5
 
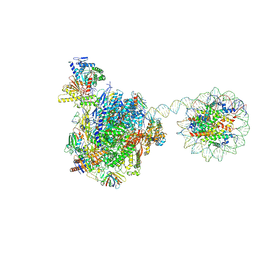 | | RNA polymerase II elongation complex bound with Rad26 and Elf1, stalled at SHL(-3.5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA repair protein, DNA-directed RNA polymerase subunit, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Kinoshita, C, Saotome, M, Kagawa, W, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (6.95 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
8H0V
 
 | | RNA polymerase II transcribing a chromatosome (type I) | | Descriptor: | DNA (261-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hirano, R, Ehara, H, Tomoya, K, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2022-09-30 | | Release date: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of RNA polymerase II transcription on the chromatosome containing linker histone H1.
Nat Commun, 13, 2022
|
|
8H0W
 
 | | RNA polymerase II transcribing a chromatosome (type II) | | Descriptor: | DNA (261-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hirano, R, Ehara, H, Tomoya, K, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2022-09-30 | | Release date: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of RNA polymerase II transcription on the chromatosome containing linker histone H1.
Nat Commun, 13, 2022
|
|
