3BDJ
 
 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with a Covalently Bound Oxipurinol Inhibitor | | Descriptor: | CALCIUM ION, CARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F, Nishino, T. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of inhibition of xanthine oxidoreductase by allopurinol: crystal structure of reduced bovine milk xanthine oxidoreductase bound with oxipurinol.
Nucleosides Nucleotides Nucleic Acids, 27, 2008
|
|
6BK9
 
 | | Crystal Structure of Squid Arrestin | | Descriptor: | CHLORIDE ION, Visual arrestin | | Authors: | Eger, B.T, Bandyopadhyay, A, Yedidi, R.S, Ernst, O.P. | | Deposit date: | 2017-11-08 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.00005579 Å) | | Cite: | A Novel Polar Core and Weakly Fixed C-Tail in Squid Arrestin Provide New Insight into Interaction with Rhodopsin.
J. Mol. Biol., 430, 2018
|
|
3UNC
 
 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase to 1.65A Resolution | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
3UNI
 
 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with NADH Bound | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-HYDROXYBENZOIC ACID, CALCIUM ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
3UNA
 
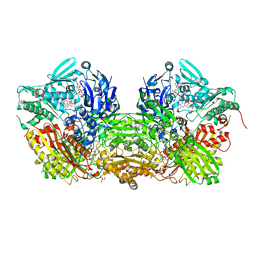 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with NAD Bound | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
6NWE
 
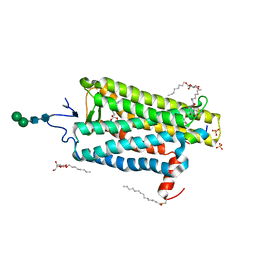 | |
6PEL
 
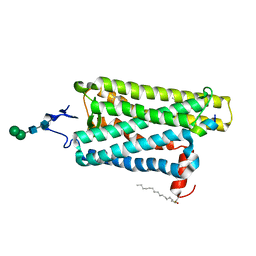 | |
6PGS
 
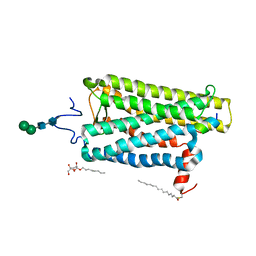 | |
6PH7
 
 | | Crystal structure of bovine opsin with nerol bound | | Descriptor: | (2Z)-3,7-dimethylocta-2,6-dien-1-ol, G protein CT2 peptide, PALMITIC ACID, ... | | Authors: | Eger, B.T, Morizumi, T, Ernst, O.P. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Odorant-binding site in visual opsin
To Be Published
|
|
4VGC
 
 | | GAMMA-CHYMOTRYPSIN D-NAPHTHYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | D-1-NAPHTHYL-2-ACETAMIDO-ETHANE BORONIC ACID, GAMMA CHYMOTRYPSIN, SULFATE ION | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
1AV7
 
 | | SUBTILISIN CARLSBERG L-NAPHTHYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-29 | | Release date: | 1998-04-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
1AVT
 
 | | SUBTILISIN CARLSBERG D-PARA-CHLOROPHENYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-19 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
1N5X
 
 | | Xanthine Dehydrogenase from Bovine Milk with Inhibitor TEI-6720 Bound | | Descriptor: | 2-(3-CYANO-4-ISOBUTOXY-PHENYL)-4-METHYL-5-THIAZOLE-CARBOXYLIC ACID, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K, Eger, B.T, Nishino, T, Kondo, S, Pai, E.F, Nishino, T. | | Deposit date: | 2002-11-07 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An Extremely Potent Inhibitor of Xanthine Oxidoreductase: Crystal Structure of the Enzyme-Inhibitor Complex and Mechanism of Inhibition
J.BIOL.CHEM., 278, 2003
|
|
3VGC
 
 | | GAMMA-CHYMOTRYPSIN L-NAPHTHYL-1-ACETAMIDO BORONIC ACID ACID INHIBITOR COMPLEX | | Descriptor: | GAMMA CHYMOTRYPSIN, L-1-NAPHTHYL-2-ACETAMIDO-ETHANE BORONIC ACID, SULFATE ION | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
3VSB
 
 | | SUBTILISIN CARLSBERG D-NAPHTHYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
5ITE
 
 | | 2.2-Angstrom in meso crystal structure of Haloquadratum Walsbyi Bacteriorhodopsin (HwBR) from Octylglucoside (OG) Detergent Micelles | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin-I, ... | | Authors: | Broecker, J, Eger, B.T, Ernst, O.P. | | Deposit date: | 2016-03-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Crystallogenesis of Membrane Proteins Mediated by Polymer-Bounded Lipid Nanodiscs.
Structure, 25, 2017
|
|
5ITC
 
 | | 2.2-Angstrom in meso crystal structure of Haloquadratum Walsbyi Bacteriorhodopsin (HwBR) from Styrene Maleic Acid (SMA) Polymer Nanodiscs | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin-I, ... | | Authors: | Broecker, J, Eger, B.T, Ernst, O.P. | | Deposit date: | 2016-03-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystallogenesis of Membrane Proteins Mediated by Polymer-Bounded Lipid Nanodiscs.
Structure, 25, 2017
|
|
5JOM
 
 | | X-ray structure of CO-bound sperm whale myoglobin using a fixed target crystallography chip | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Oghbaey, S, Sarracini, A, Ginn, H.M, Pare-Labrosse, O, Kuo, A, Marx, A, Epp, S.W, Sherrell, D.A, Eger, B.T, Zhong, Y, Loch, R, Mariani, V, Alonso-Mori, R, Nelson, S, Lemke, H.T, Owen, R.L, Pearson, A.R, Stuart, D.I, Ernst, O.P, Mueller-Werkmeister, H.M, Miller, R.J.D. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fixed target combined with spectral mapping: approaching 100% hit rates for serial crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7F1W
 
 | | X-ray crystal structure of visual arrestin complexed with inositol hexaphosphate | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, S-arrestin | | Authors: | Kang, M, Jang, K, Eger, B.T, Ernst, O.P, Choe, H.W, Kim, Y.J. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7F1X
 
 | | X-ray crystal structure of visual arrestin complexed with inositol 1,4,5-triphosphate | | Descriptor: | 1,2-ETHANEDIOL, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PENTANEDIAL, ... | | Authors: | Jang, K, Kang, M, Eger, B.T, Choe, H.W, Ernst, O.P, Kim, Y.J. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
4YTY
 
 | | Structure of rat xanthine oxidoreductase, C535A/C992R/C1324S, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase.
Febs J., 282, 2015
|
|
4YSW
 
 | | Structure of rat xanthine oxidoreductase, C-terminal deletion protein variant, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F. | | Deposit date: | 2015-03-17 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase.
Febs J., 282, 2015
|
|
4YTZ
 
 | | Rat xanthine oxidoreductase, C-terminal deletion protein variant, crystal grown without dithiothreitol | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase
Febs J., 282, 2015
|
|
1V97
 
 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase FYX-051 bound form | | Descriptor: | 4-(5-PYRIDIN-4-YL-1H-1,2,4-TRIAZOL-3-YL)PYRIDINE-2-CARBONITRILE, ACETIC ACID, CALCIUM ION, ... | | Authors: | Okamoto, K, Matsumoto, K, Hille, R, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2004-01-21 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The crystal structure of xanthine oxidoreductase during catalysis: Implications for reaction mechanism and enzyme inhibition.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5DKP
 
 | | Crystal Structure of N. meningitidis ClpP in complex with agonist ADEP A54556. | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, POTASSIUM ION, SODIUM ION, ... | | Authors: | Goodreid, J.D, Janetzko, J, Santa Maria Jr, J.P, Wong, K, Leung, E, Eger, B.T, Bryson, S, Pai, E.F, Gray-Owen, S.D, Walker, S, Houry, W.A, Batey, R.A. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Development and Characterization of Potent Cyclic Acyldepsipeptide Analogues with Increased Antimicrobial Activity.
J.Med.Chem., 59, 2016
|
|
