2HOK
 
 | |
2HOM
 
 | |
2HOL
 
 | |
2HOJ
 
 | |
2HOP
 
 | |
2HOO
 
 | |
6AQ3
 
 | |
6AY1
 
 | |
6AMZ
 
 | | Crystal structure of a domain swapped 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase from Acinetobacter baumannii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase, CALCIUM ION | | Authors: | Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2017-08-11 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a domain swapped 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase from Acinetobacter baumannii
To Be Published
|
|
3TE8
 
 | |
5TQJ
 
 | |
3IEF
 
 | |
3OT0
 
 | |
4WTD
 
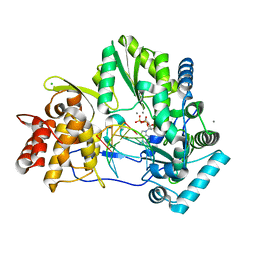 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS AND DELTA8 BETA HAIRPIN LOOP DELETION IN COMPLEX WITH ADP, MN2+ AND SYMMETRICAL PRIMER TEMPLATE 5'-AUAAAUUU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Edwards, T.E, Fox III, D, Appleby, T.C. | | Deposit date: | 2014-10-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTF
 
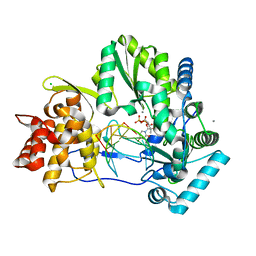 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS AND DELTA8 BETA HAIRPIN LOOP DELETION IN COMPLEX WITH GS-639475, MN2+ AND SYMMETRICAL PRIMER TEMPLATE 5'-CAAAAUUU | | Descriptor: | 2'-C-methyluridine 5'-(trihydrogen diphosphate), CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Edwards, T.E, Appleby, T.C, Mcgrath, M.E. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTJ
 
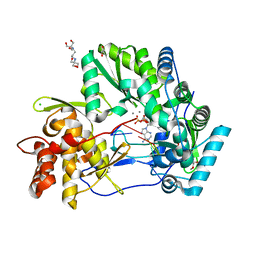 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-AUCC, RNA PRIMER 5'-PGG, MN2+, AND ADP | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTM
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-UAGG, RNA PRIMER 5'-PCC, MN2+, AND UDP | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, RNA PRIMER CC, ... | | Authors: | Edwards, T.E, Appleby, T.C, Fox III, D. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WT9
 
 | |
4WTA
 
 | |
4WTC
 
 | |
4WTE
 
 | |
4WTG
 
 | |
4WTI
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-ACGG, RNA PRIMER 5'-PCC, MN2+, AND GDP | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTK
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-AGCC, RNA PRIMER 5'-PGG, MN2+, AND CDP | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTL
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-UACC, RNA PRIMER 5'-PGG, MN2+, AND UDP | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
