5E0A
 
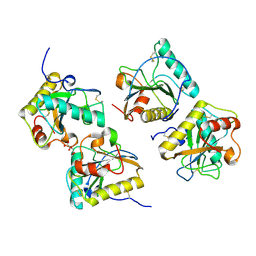 | | Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A
To Be Published
|
|
3NNO
 
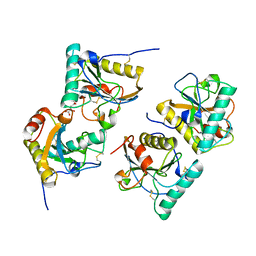 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with Alpha-Rhamnose at 2.9 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1, alpha-L-rhamnopyranose | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with Alpha-Rhamnose at 2.9 A resolution
To be Published
|
|
3QS0
 
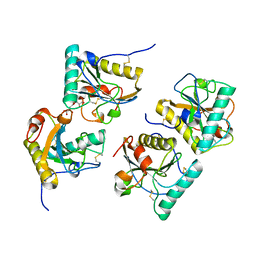 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with a bound N-acetylglucosamine in the diffusion channel at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-02-19 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with a bound N-acetylglucosamine in the diffusion channel AT 2.5 A resolution
To be Published
|
|
3RKE
 
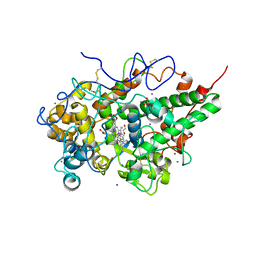 | | Crystal Structure of goat Lactoperoxidase complexed with a tightly bound inhibitor, 4-aminophenyl-4H-imidazole-1-yl methanone at 2.3 A resolution | | Descriptor: | (4-aminophenyl)-imidazol-1-yl-methanone, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dube, D, Singh, R.P, Sinha, M, Singh, A.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of goat Lactoperoxidase complexed with a tightly bound inhibitor, 4-aminophenyl-4H-imidazole-1-yl methanone at 2.3 A resolution
TO BE PUBLISHED
|
|
3TRU
 
 | | Crystal structure of the complex of peptidoglycan recognition protein with cellular metabolite chorismate at 3.2 A resolution | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-09-10 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein with cellular metabolite chorismate at 3.2 A resolution
To be Published
|
|
4FNN
 
 | | Crystal structure of the complex of CPGRP-S with stearic acid at 2.2 A RESOLUTION | | Descriptor: | Peptidoglycan recognition protein 1, STEARIC ACID | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site.
Arch.Biochem.Biophys., 529, 2013
|
|
3UIL
 
 | | Crystal Structure of the complex of PGRP-S with lauric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, LAURIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-05 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
2VU5
 
 | | Crystal structure of Pndk from Bacillus anthracis | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Misra, G, Aggarwal, A, Dube, D, Zaman, M.S, Singh, Y, Ramachandran, R. | | Deposit date: | 2008-05-21 | | Release date: | 2009-03-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Bacillus Anthracis Nucleoside Diphosphate Kinase and its Characterization Reveals an Enzyme Adapted to Perform Under Stress Conditions.
Proteins, 76, 2009
|
|
5E0B
 
 | | Crystal structure of the complex of Peptidoglycan recognition protein PGRP-S with N-Acetyl Muramic acid at 2.6 A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, N-acetyl-beta-muramic acid, ... | | Authors: | Sharma, P, Yamini, S, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of Peptidoglycan recognition protein PGRP-S with N-Acetyl Muramic acid at 2.6 A resolution
To Be Published
|
|
4GF9
 
 | | Structural insights into the dual strategy of recognition of peptidoglycan recognition protein, PGRP-S: ternary complex of PGRP-S with LPS and fatty acid | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Yadav, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-03 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the dual strategy of recognition by peptidoglycan recognition protein, PGRP-S: structure of the ternary complex of PGRP-S with lipopolysaccharide and stearic acid.
Plos One, 8, 2013
|
|
3M7S
 
 | | Crystal structure of the complex of xylanase GH-11 and alpha amylase inhibitor protein with cellobiose at 2.4 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, S, Dube, D, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
4LL2
 
 | | Crystal structure of plant lectin with two metal binding sites from cicer arietinum at 2.6 angstrom resolution | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Lectin, ... | | Authors: | Kumar, S, Dube, D, Bhushan, A, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-09 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure plant lectinwith two metal binding sites from cicer arietinum at 2.6 angstrom resolution
TO BE PUBLISHED
|
|
4N6P
 
 | | Crystal Structure of C-lobe of Bovine lactoferrin complexed with meclofenamic acid at 1.4 A resolution | | Descriptor: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gautam, L, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-10-14 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structre of C-LOBE OF BOVINE LACTOFERRIN COMPLEXED WITH MECLOFENAMIC ACID AT 1.4 A RESOLUTION
TO BE PUBLISHED
|
|
4NED
 
 | | Crystal STRUCTURE OF C-LOBE OF BOVINE LACTOFERRIN COMPLEXED WITH FENOPROFEN AT 2.1 ANGSTROM RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Gautam, L, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-10-29 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal STRUCTURE OF C-LOBE OF BOVINE LACTOFERRIN COMPLEXED WITH FENOPROFEN AT 2.1 ANGSTROM RESOLUTION
To be Published
|
|
4OLJ
 
 | | Crystal structure of Arg119Gln mutant of Peptidyl-tRNA Hydrolase from Acinetobacter Baumannii at 1.49 A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Peptidyl-tRNA hydrolase | | Authors: | Sikarwar, J, Dube, D, Kaushik, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-01-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of Arg119Gln mutant of Peptidyl-tRNA hydrolase from Acinetobacter Baumannii at 1.49 A resolution
to be published
|
|
4OEI
 
 | | Crystal structure of plant lectin from Cicer arietinum at 2.6 angstrom resolution | | Descriptor: | Lectin, MAGNESIUM ION, SULFATE ION | | Authors: | Kumar, S, Dube, D, Bhushan, A, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2014-01-13 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of plant lectin from Cicer arietinum at
2.6 angstrom resolution
TO BE PUBLISHED
|
|
3NG4
 
 | | Ternary complex of peptidoglycan recognition protein (PGRP-S) with Maltose and N-Acetylglucosamine at 1.7 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Multiligand specificity of pathogen-associated molecular pattern-binding site in peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
3NW3
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with the PGN Fragment at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, ALANINE, D-GLUTAMINE, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multiligand specificity of pathogen-associated molecular pattern-binding site in peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
3O4K
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) and lipoteichoic acid at 2.1 A resolution | | Descriptor: | (2S)-1-({3-O-[2-(acetylamino)-4-amino-2,4,6-trideoxy-beta-D-galactopyranosyl]-alpha-D-glucopyranosyl}oxy)-3-(heptanoyloxy)propan-2-yl (7Z)-pentadec-7-enoate, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural basis of recognition of pathogen-associated molecular patterns and inhibition of proinflammatory cytokines by camel peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
3RT4
 
 | | Structural Basis of Recognition of Pathogen-associated Molecular Patterns and Inhibition of Proinflammatory Cytokines by Camel Peptidoglycan Recognition Protein | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Sharma, P, Dube, D, Singh, A, Mishra, B, Singh, N, Sinha, M, Dey, S, Kaur, P, Mitra, D.K, Sharma, S, Singh, T.P. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Recognition of Pathogen-associated Molecular Patterns and Inhibition of Proinflammatory Cytokines by Camel Peptidoglycan Recognition Protein.
J.Biol.Chem., 286, 2011
|
|
3OGX
 
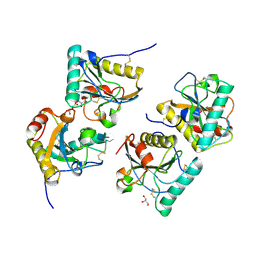 | | Crystal structure of the complex of Peptidoglycan Recognition protein (PGRP-s) with Heparin-Dissacharide at 2.8 A resolution | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of heparin binding to camel peptidoglycan recognition protein-S
Int J Biochem Mol Biol, 3, 2012
|
|
3T2V
 
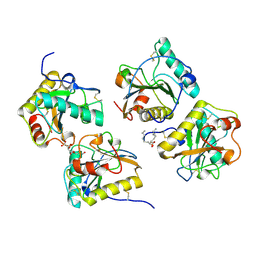 | | Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with mycolic acid at 2.5 A resolution | | Descriptor: | (2S,3R)-2-hexyl-3-hydroxynonanoic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3T39
 
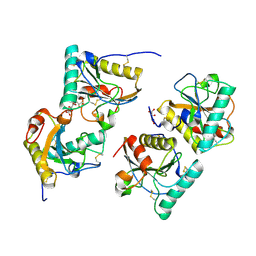 | | Crystal structure of the complex of camel peptidoglycan recognition protein(CPGRP-S) with a mycobacterium metabolite shikimate at 2.7 A resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with a mycobacterium metabolite shikimate at 2.7 A resolution
To be Published
|
|
3HU7
 
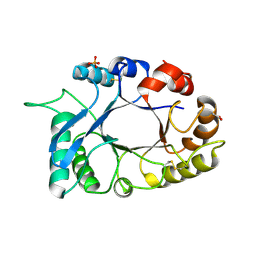 | | Structural characterization and binding studies of a plant pathogenesis related protein heamanthin from haemanthus multiflorus reveal its dual inhibitory effects against xylanase and alpha-amylase | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Kumar, S, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-06-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
3MU7
 
 | | Crystal structure of the xylanase and alpha-amylase inhibitor protein (XAIP-II) from scadoxus multiflorus at 1.2 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, xylanase and alpha-amylase inhibitor protein | | Authors: | Kumar, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Modulation of inhibitory activity of xylanase-alpha-amylase inhibitor protein (XAIP): binding studies and crystal structure determination of XAIP-II from Scadoxus multiflorus at 1.2 A resolution.
Bmc Struct.Biol., 10, 2010
|
|
