1BDS
 
 | |
2BDS
 
 | |
1E3Y
 
 | |
1E41
 
 | |
1VCA
 
 | | CRYSTAL STRUCTURE OF AN INTEGRIN-BINDING FRAGMENT OF VASCULAR CELL ADHESION MOLECULE-1 AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN VASCULAR CELL ADHESION MOLECULE-1 | | Authors: | Jones, E.Y, Harlos, K, Bottomley, M.J, Robinson, R.C, Driscoll, P.C, Edwards, R.M, Clements, J.M, Dudgeon, T.J, Stuart, D.I. | | Deposit date: | 1995-03-21 | | Release date: | 1995-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 A resolution.
Nature, 373, 1995
|
|
1BFI
 
 | | SOLUTION STRUCTURE OF THE C-TERMINAL SH2 DOMAIN OF THE P85ALPHA REGULATORY SUBUNIT OF PHOSPHOINOSITIDE 3-KINASE, NMR, 30 STRUCTURES | | Descriptor: | P85 ALPHA | | Authors: | Siegal, G, Davis, B, Kristensen, S.M, Sankar, A, Linacre, J, Stein, R.C, Panayotou, G, Waterfield, M.D, Driscoll, P.C. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the p85 alpha regulatory subunit of phosphoinositide 3-kinase.
J.Mol.Biol., 276, 1998
|
|
1BFJ
 
 | | SOLUTION STRUCTURE OF THE C-TERMINAL SH2 DOMAIN OF THE P85ALPHA REGULATORY SUBUNIT OF PHOSPHOINOSITIDE 3-KINASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | P85 ALPHA | | Authors: | Siegal, G, Davis, B, Kristensen, S.M, Sankar, A, Linacre, J, Stein, R.C, Panayotou, G, Waterfield, M.D, Driscoll, P.C. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the p85 alpha regulatory subunit of phosphoinositide 3-kinase.
J.Mol.Biol., 276, 1998
|
|
1TPG
 
 | | F1-G MODULE PAIR RESIDUES 1-91 (C83S) OF TISSUE-TYPE PLASMINOGEN ACTIVATOR (T-PA) (NMR, 298K, PH2.95, REPRESENTATIVE STRUCTURE) | | Descriptor: | T-PLASMINOGEN ACTIVATOR F1-G | | Authors: | Smith, B.O, Downing, A.K, Driscoll, P.C, Dudgeon, T.J, Campbell, I.D. | | Deposit date: | 1995-06-14 | | Release date: | 1995-09-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure and backbone dynamics of the fibronectin type I and epidermal growth factor-like pair of modules of tissue-type plasminogen activator.
Structure, 3, 1995
|
|
2AR5
 
 | |
2BYE
 
 | | NMR solution structure of phospholipase c epsilon RA 1 domain | | Descriptor: | PHOSPHOLIPASE C, EPSILON 1 | | Authors: | Bunney, T.D, Harris, R, Gandarillas, N.L, Josephs, M.B, Roe, S.M, Paterson, H.F, Rodrigues-Lima, F, Esposito, D, Gieschik, P, Pearl, L.H, Driscoll, P.C, Katan, M. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon.
Mol.Cell, 21, 2006
|
|
2BYF
 
 | | NMR solution structure of phospholipase c epsilon RA 2 domain | | Descriptor: | PHOSPHOLIPASE C, EPSILON 1 | | Authors: | Bunney, T.D, Harris, R, Gandarillas, N.L, Josephs, M.B, Roe, S.M, Paterson, H.F, Rodrigues-Lima, F, Esposito, D, Gieschik, P, Pearl, L.H, Driscoll, P.C, Katan, M. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon.
Mol.Cell, 21, 2006
|
|
1NHN
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
NUCLEIC ACIDS MOL.BIOL., 9, 1995
|
|
1NHM
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
NUCLEIC ACIDS MOL.BIOL., 9, 1995
|
|
1HSN
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | BETA-MERCAPTOETHANOL, HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
To be Published
|
|
1HSM
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | BETA-MERCAPTOETHANOL, HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
To be Published
|
|
1TPM
 
 | | SOLUTION STRUCTURE OF THE FIBRIN BINDING FINGER DOMAIN OF TISSUE-TYPE PLASMINOGEN ACTIVATOR DETERMINED BY 1H NUCLEAR MAGNETIC RESONANCE | | Descriptor: | TISSUE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Downing, A.K, Driscoll, P.C, Harvey, T.S, Dudgeon, T.J, Smith, B.O, Baron, M, Campbell, I.D. | | Deposit date: | 1993-05-26 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fibrin binding finger domain of tissue-type plasminogen activator determined by 1H nuclear magnetic resonance.
J.Mol.Biol., 225, 1992
|
|
1TPN
 
 | | SOLUTION STRUCTURE OF THE FIBRIN BINDING FINGER DOMAIN OF TISSUE-TYPE PLASMINOGEN ACTIVATOR DETERMINED BY 1H NUCLEAR MAGNETIC RESONANCE | | Descriptor: | TISSUE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Downing, A.K, Driscoll, P.C, Harvey, T.S, Dudgeon, T.J, Smith, B.O, Baron, M, Campbell, I.D. | | Deposit date: | 1993-05-26 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fibrin binding finger domain of tissue-type plasminogen activator determined by 1H nuclear magnetic resonance.
J.Mol.Biol., 225, 1992
|
|
2OFQ
 
 | | NMR Solution Structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system | | Descriptor: | TraN, TraO | | Authors: | Harris, R, Bayliss, R, Driscoll, P.C, Waksman, G. | | Deposit date: | 2007-01-04 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PNI
 
 | | SOLUTION STRUCTURE AND LIGAND-BINDING SITE OF THE SH3 DOMAIN OF THE P85ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL 3-KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Booker, G.W, Gout, I, Downing, A.K, Driscoll, P.C, Boyd, J, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1993-07-19 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and ligand-binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3-kinase.
Cell(Cambridge,Mass.), 73, 1993
|
|
2REA
 
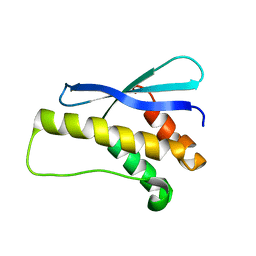 | |
2RED
 
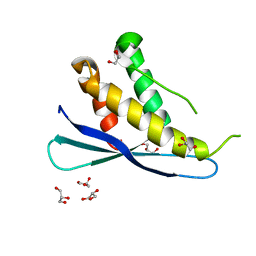 | | Crystal structures of C2ALPHA-PI3 kinase PX-domain domain indicate conformational change associated with ligand binding. | | Descriptor: | GLYCEROL, Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide | | Authors: | Parkinson, G.N, Vines, D, Driscoll, P.C, Djordjevic, S. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of PI3K-C2alpha PX domain indicate conformational change associated with ligand binding
Bmc Struct.Biol., 8, 2008
|
|
1ERH
 
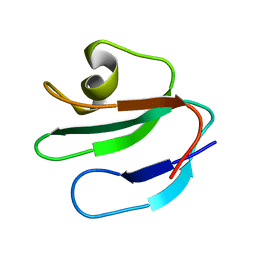 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE EXTRACELLULAR REGION OF THE COMPLEMENT REGULATORY PROTEIN, CD59, A NEW CELL SURFACE PROTEIN DOMAIN RELATED TO NEUROTOXINS | | Descriptor: | CD59 | | Authors: | Kieffer, B, Driscoll, P.C, Campbell, I.D, Willis, A.C, Van Der Merwe, P.A, Davis, S.J. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the extracellular region of the complement regulatory protein CD59, a new cell-surface protein domain related to snake venom neurotoxins.
Biochemistry, 33, 1994
|
|
1ERG
 
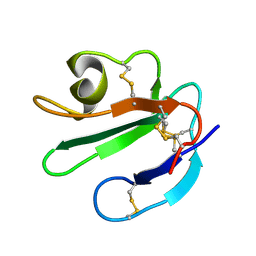 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE EXTRACELLULAR REGION OF THE COMPLEMENT REGULATORY PROTEIN, CD59, A NEW CELL SURFACE PROTEIN DOMAIN RELATED TO NEUROTOXINS | | Descriptor: | CD59 | | Authors: | Kieffer, B, Driscoll, P.C, Campbell, I.D, Willis, A.C, Van Der Merwe, P.A, Davis, S.J. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the extracellular region of the complement regulatory protein CD59, a new cell-surface protein domain related to snake venom neurotoxins.
Biochemistry, 33, 1994
|
|
2JVE
 
 | | Solution structure of the extracellular domain of Prod1, a protein implicated in proximodistal identity during amphibian limb regeneration | | Descriptor: | Prod 1 | | Authors: | Garza-Garcia, A, Harris, R, Esposito, D, Driscoll, P.C. | | Deposit date: | 2007-09-19 | | Release date: | 2008-09-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure and phylogenetics of Prod1, a member of the three-finger protein superfamily implicated in salamander limb regeneration.
Plos One, 4, 2009
|
|
2K2J
 
 | | NMR solution structure of the split PH domain from Phospholipase C gamma 2 | | Descriptor: | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-2 | | Authors: | Harris, R, Bunney, T.D, Katan, M, Driscoll, P.C. | | Deposit date: | 2008-04-02 | | Release date: | 2008-09-09 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Rac Regulates Its Effector Phospholipase C{gamma}2 through Interaction with a Split Pleckstrin Homology Domain.
J.Biol.Chem., 283, 2008
|
|
