3L94
 
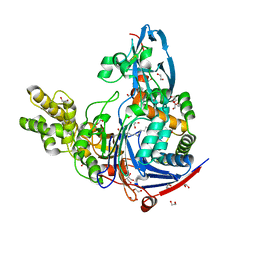 | | Structure of PvdQ covalently acylated with myristate | | Descriptor: | 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase pvdQ subunit alpha, Acyl-homoserine lactone acylase pvdQ subunit beta, ... | | Authors: | Drake, E.J, Gulick, A.M. | | Deposit date: | 2010-01-04 | | Release date: | 2011-01-26 | | Last modified: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization and high-throughput screening of inhibitors of PvdQ, an NTN hydrolase involved in pyoverdine synthesis.
Acs Chem.Biol., 6, 2011
|
|
4ZXI
 
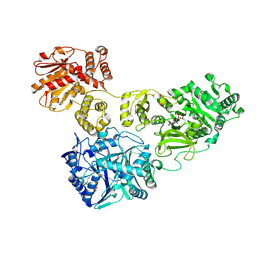 | | Crystal Structure of holo-AB3403 a four domain nonribosomal peptide synthetase bound to AMP and Glycine | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 4'-PHOSPHOPANTETHEINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Drake, E.J, Miller, B.R, Allen, C.L, Gulick, A.M. | | Deposit date: | 2015-05-20 | | Release date: | 2015-12-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of two distinct conformations of holo-non-ribosomal peptide synthetases.
Nature, 529, 2016
|
|
4ZXH
 
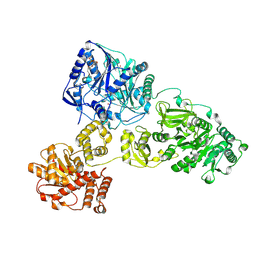 | | Crystal Structure of holo-AB3403 a four domain nonribosomal peptide synthetase from Acinetobacter Baumanii | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4'-PHOSPHOPANTETHEINE, ABBFA_003403, ... | | Authors: | Drake, E.J, Miller, B.R, Allen, C.L, Gulick, A.M. | | Deposit date: | 2015-05-20 | | Release date: | 2015-12-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of two distinct conformations of holo-non-ribosomal peptide synthetases.
Nature, 529, 2016
|
|
2FQ1
 
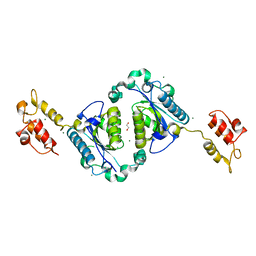 | | Crystal structure of the two-domain non-ribosomal peptide synthetase EntB containing isochorismate lyase and aryl-carrier protein domains | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Isochorismatase, ... | | Authors: | Drake, E.J, Nicolai, D.A, Gulick, A.M. | | Deposit date: | 2006-01-17 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the EntB multidomain nonribosomal peptide synthetase and functional analysis of its interaction with the EntE adenylation domain.
Chem.Biol., 13, 2006
|
|
3L91
 
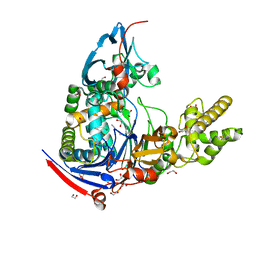 | | Structure of Pseudomonas aerugionsa PvdQ bound to octanoate | | Descriptor: | 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase pvdQ subunit alpha, Acyl-homoserine lactone acylase pvdQ subunit beta, ... | | Authors: | Drake, E.J, Gulick, A.M. | | Deposit date: | 2010-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural characterization and high-throughput screening of inhibitors of PvdQ, an NTN hydrolase involved in pyoverdine synthesis.
Acs Chem.Biol., 6, 2011
|
|
3O83
 
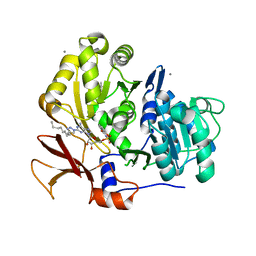 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 2-(4-n-dodecyl-1,2,3-triazol-1-yl)-5'-O-[N-(2-hydroxybenzoyl)sulfamoyl]adenosine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-dodecyl-1H-1,2,3-triazol-1-yl)-5'-O-{[(2-hydroxyphenyl)carbonyl]sulfamoyl}adenosine, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
3O84
 
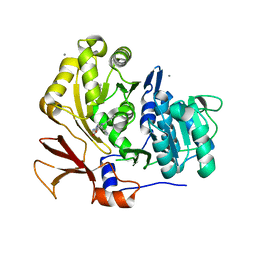 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
3O82
 
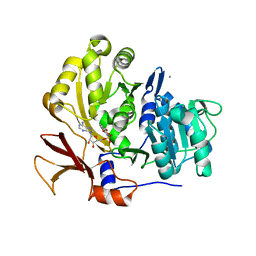 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 5'-O-[N-(2,3-dihydroxybenzoyl)sulfamoyl] adenosine | | Descriptor: | 5'-O-{[(2,3-dihydroxyphenyl)carbonyl]sulfamoyl}adenosine, CALCIUM ION, Peptide arylation enzyme | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
5I90
 
 | | Crystal Structure of PvdN from Pseudomonas Aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, PYRIDOXAL-5'-PHOSPHATE, PvdN | | Authors: | Drake, E.J, Gulick, A.M. | | Deposit date: | 2016-02-19 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | 1.2 angstrom resolution crystal structure of the periplasmic aminotransferase PvdN from Pseudomonas aeruginosa.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5T3D
 
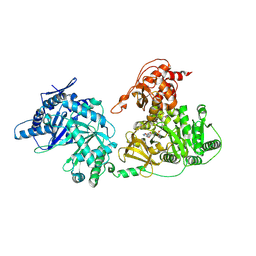 | | Crystal structure of holo-EntF a nonribosomal peptide synthetase in the thioester-forming conformation | | Descriptor: | 5'-({[(2R,3S)-3-amino-4-hydroxy-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}butyl]sulfonyl}amino)-5'-deoxyadenosine, Enterobactin synthase component F | | Authors: | Miller, B.R, Drake, E.J, Sundlov, J.A, Gulick, A.M. | | Deposit date: | 2016-08-25 | | Release date: | 2016-09-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of two distinct conformations of holo-non-ribosomal peptide synthetases.
Nature, 529, 2016
|
|
3E59
 
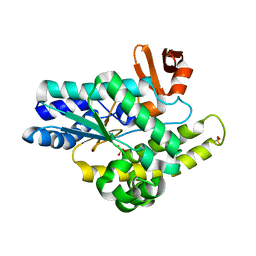 | |
3EAT
 
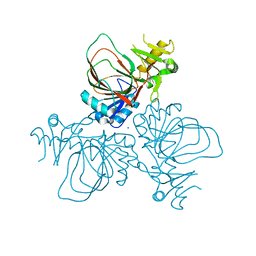 | |
2PST
 
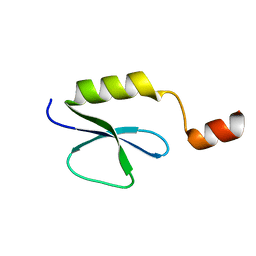 | |
3SRC
 
 | | Structure of Pseudomonas aeruginosa PvdQ bound to NS2028 | | Descriptor: | 1,2-ETHANEDIOL, 8-bromo-4H-[1,2,4]oxadiazolo[3,4-c][1,4]benzoxazin-1-one, Acyl-homoserine lactone acylase pvdQ | | Authors: | Gulick, A.M, Drake, E.J. | | Deposit date: | 2011-07-07 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization and High-Throughput Screening of Inhibitors of PvdQ, an NTN Hydrolase Involved in Pyoverdine Synthesis.
Acs Chem.Biol., 6, 2011
|
|
3SRA
 
 | | Structure of Pseudomonas aerugionsa PvdQ covalently acylated with myristic acid from PVDIq | | Descriptor: | 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase PvdQ subunit alpha, Acyl-homoserine lactone acylase PvdQ subunit beta, ... | | Authors: | Gulick, A.M, Drake, E.J. | | Deposit date: | 2011-07-07 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization and High-Throughput Screening of Inhibitors of PvdQ, an NTN Hydrolase Involved in Pyoverdine Synthesis.
Acs Chem.Biol., 6, 2011
|
|
3SRB
 
 | | Structure of Pseudomonas aeruginosa PvdQ bound to SMER28 | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-N-(prop-2-en-1-yl)quinazolin-4-amine, Acyl-homoserine lactone acylase pvdQ, ... | | Authors: | Gulick, A.M, Drake, E.J. | | Deposit date: | 2011-07-07 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Characterization and High-Throughput Screening of Inhibitors of PvdQ, an NTN Hydrolase Involved in Pyoverdine Synthesis.
Acs Chem.Biol., 6, 2011
|
|
4K2G
 
 | | Structure of Pseudomonas aeruginosa PvdQ bound to BRD-A33442372 | | Descriptor: | (2S)-(4-fluorophenyl)[6-(trifluoromethyl)pyridin-2-yl]ethanenitrile, 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase PvdQ | | Authors: | Drake, E.J, Wurst, J.M, Theriault, J.R, Munoz, B, Gulick, A.M. | | Deposit date: | 2013-04-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Inhibitors of PvdQ, an Enzyme Involved in the Synthesis of the Siderophore Pyoverdine.
Acs Chem.Biol., 9, 2014
|
|
4K2F
 
 | | Structure of Pseudomonas aeruginosa PvdQ bound to BRD-A08522488 | | Descriptor: | (2S)-(4-chlorophenyl)(6-chloropyridin-2-yl)ethanenitrile, 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase PvdQ | | Authors: | Drake, E.J, Wurst, J.M, Theriault, J.R, Munoz, B, Gulick, A.M. | | Deposit date: | 2013-04-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Identification of Inhibitors of PvdQ, an Enzyme Involved in the Synthesis of the Siderophore Pyoverdine.
Acs Chem.Biol., 9, 2014
|
|
5I3F
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3H
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, POTASSIUM ION, Triosephosphate isomerase, ... | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3J
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | SODIUM ION, Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3I
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3K
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, SODIUM ION, Triosephosphate isomerase, ... | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3G
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5JM7
 
 | |
