3KO1
 
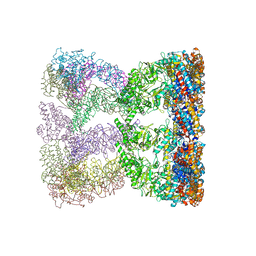 | | Cystal structure of thermosome from Acidianus tengchongensis strain S5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin | | Authors: | Huo, Y, Zhang, K, Hu, Z, Wang, L, Zhai, Y, Zhou, Q, Lander, G, He, Y, Zhu, J, Xu, W, Dong, Z, Sun, F. | | Deposit date: | 2009-11-12 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of group II chaperonin in the open state.
Structure, 18, 2010
|
|
3KYS
 
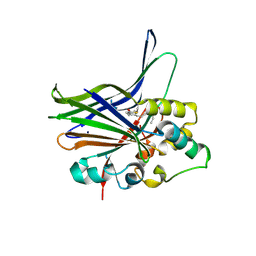 | | Crystal structure of human YAP and TEAD complex | | Descriptor: | 65 kDa Yes-associated protein, Transcriptional enhancer factor TEF-1 | | Authors: | Li, Z, Zhao, B, Wang, P, Chen, F, Dong, Z, Yang, H, Guan, K.L, Xu, Y. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-23 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the YAP and TEAD complex
Genes Dev., 24, 2010
|
|
3V3V
 
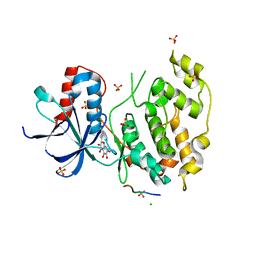 | | Structural and functional analysis of quercetagetin, a natural JNK1 inhibitor | | Descriptor: | 3,5,6,7-TETRAHYDROXY-2-(3,4-DIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, C-Jun-amino-terminal kinase-interacting protein 1, CHLORIDE ION, ... | | Authors: | Baek, S, Kang, N.J, Popowicz, G.M, Arciniega, M, Jung, S.K, Byun, S, Song, N.R, Heo, Y.S, Kim, B.Y, Lee, H.J, Holak, T.A, Augustin, M, Bode, A.M, Huber, R, Dong, Z, Lee, K.W. | | Deposit date: | 2011-12-14 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Analysis of the Natural JNK1 Inhibitor Quercetagetin.
J.Mol.Biol., 425, 2013
|
|
8YCM
 
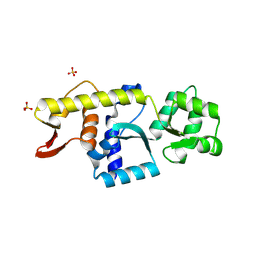 | | Monomeric Human STK19 | | Descriptor: | Isoform 4 of Inactive serine/threonine-protein kinase 19, SULFATE ION | | Authors: | Li, J, Ma, X, Dong, Z. | | Deposit date: | 2024-02-18 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Monomeric human STK19
To Be Published
|
|
2QR8
 
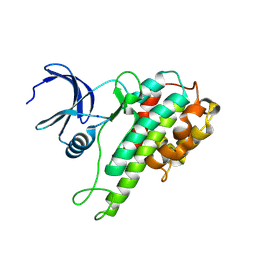 | |
2QR7
 
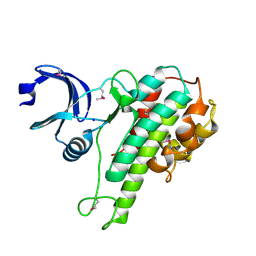 | |
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUR
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUT
 
 | | Crystal structure of LSD2-NPAC | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GU1
 
 | | Crystal structure of LSD2 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.939 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUU
 
 | | Crystal structure of LSD2-NPAC with tranylcypromine | | Descriptor: | Lysine-specific histone demethylase 1B, Putative oxidoreductase GLYR1, ZINC ION, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
3KN6
 
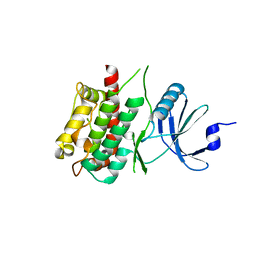 | |
3KN5
 
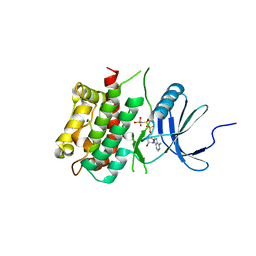 | |
4GU0
 
 | | Crystal structure of LSD2 with H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Yang, H, Dong, Z, Fang, J, Zhu, T, Gong, W, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural insight into substrate recognition by histone demethylase LSD2/KDM1b
Cell Res., 23, 2013
|
|
4HSU
 
 | | Crystal structure of LSD2-NPAC with H3(1-26)in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Xu, Y. | | Deposit date: | 2012-10-30 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural insight into substrate recognition by histone demethylase LSD2/KDM1b.
Cell Res., 23, 2013
|
|
3OOB
 
 | | Structural and functional insights of directly targeting Pin1 by Epigallocatechin-3-gallate | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Urusova, D.V, Shim, J.-H, Kim, D.-J, Jung, S.K, Zykova, T.A, Bode, A.M, Dong, Z. | | Deposit date: | 2010-08-30 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Epigallocatechin-gallate suppresses tumorigenesis by directly targeting Pin1.
Cancer Prev Res (Phila), 4, 2011
|
|
6SLG
 
 | | HUMAN ERK2 WITH ERK1/2 INHIBITOR, AZD0364. | | Descriptor: | (6~{R})-7-[[3,4-bis(fluoranyl)phenyl]methyl]-6-(methoxymethyl)-2-[5-methyl-2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-5,6-dihydroimidazo[1,2-a]pyrazin-8-one, 1,2-ETHANEDIOL, ERK-tide, ... | | Authors: | Breed, J, Phillips, C. | | Deposit date: | 2019-08-19 | | Release date: | 2019-11-20 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a Potent and Selective Oral Inhibitor of ERK1/2 (AZD0364) That Is Efficacious in Both Monotherapy and Combination Therapy in Models of Nonsmall Cell Lung Cancer (NSCLC).
J.Med.Chem., 62, 2019
|
|
6AKW
 
 | | Crystal structure of RNA dioxygenase bound with an inhibitor | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[[2,6-bis(chloranyl)-4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]amino]benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO | | Authors: | Yang, C.-G, Huang, Y, Gan, J. | | Deposit date: | 2018-09-04 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia.
Cancer Cell, 35, 2019
|
|
4ZXT
 
 | | Complex of ERK2 with catechol | | Descriptor: | AMMONIUM ION, CATECHOL, Mitogen-activated protein kinase 1, ... | | Authors: | Kurinov, I, Malakhova, M. | | Deposit date: | 2015-05-20 | | Release date: | 2016-05-25 | | Last modified: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A natural small molecule, catechol, induces c-Myc degradation by directly targeting ERK2 in lung cancer.
Oncotarget, 7, 2016
|
|
4R9I
 
 | | Crystal structure of cysteine proteinase inhibitor Serpin18 from Bombyx mori | | Descriptor: | BETA-MERCAPTOETHANOL, CITRATE ANION, SODIUM ION, ... | | Authors: | Guo, P.C, He, H.W, Zhao, P, Xia, Q.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the unique inhibitory mechanism of the silkworm protease inhibitor serpin18
Sci Rep, 5, 2015
|
|
4RYX
 
 | | Crystal structure of RPE65 in complex with emixustat and palmitate, P6522 crystal form | | Descriptor: | (1R)-3-amino-1-[3-(cyclohexylmethoxy)phenyl]propan-1-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, FE (II) ION, ... | | Authors: | Kiser, P.D, Palczewski, K. | | Deposit date: | 2014-12-17 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular pharmacodynamics of emixustat in protection against retinal degeneration.
J.Clin.Invest., 125, 2015
|
|
4RYY
 
 | |
4RYZ
 
 | |
4N0S
 
 | | Complex of ERK2 with caffeic acid | | Descriptor: | CAFFEIC ACID, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Kurinov, I, Malakhova, M. | | Deposit date: | 2013-10-02 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7992 Å) | | Cite: | Caffeic Acid Directly Targets ERK1/2 to Attenuate Solar UV-Induced Skin Carcinogenesis.
Cancer Prev Res (Phila), 7, 2014
|
|
6WLG
 
 | | Ints3 C-terminal Domain | | Descriptor: | Integrator complex subunit 3 | | Authors: | Li, J, Ma, X.L, Banerjee, S, Dong, Z.G. | | Deposit date: | 2020-04-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.111 Å) | | Cite: | Structural basis for multifunctional roles of human Ints3 C-terminal domain.
J.Biol.Chem., 296, 2020
|
|
