2YJY
 
 | | A specific and modular binding code for cytosine recognition in PUF domains | | Descriptor: | 5'-R(*AP*UP*UP*GP*CP*AP*UP*AP*UP*AP)-3', PUMILIO HOMOLOG 1 | | Authors: | Dong, S, Wang, Y, Cassidy-Amstutz, C, Lu, G, Qiu, C, Bigler, R, Jezyk, M, Li, C, Hall, T.M.T, Wang, Z. | | Deposit date: | 2011-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Specific and Modular Binding Code for Cytosine Recognition in Pumilio/Fbf (Puf) RNA-Binding Domains.
J.Biol.Chem., 286, 2011
|
|
8WBS
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases KlCESH[L]-D48N complexed with sulfate ions | | Descriptor: | (S)-2-haloacid dehalogenase, CALCIUM ION, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBN
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant D193N | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBK
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBR
 
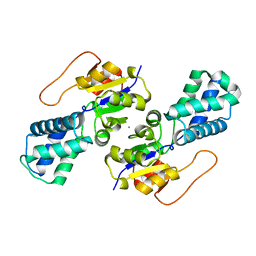 | | Crystal structure of cis-Epoxysuccinate Hydrolases KlCESH[L] | | Descriptor: | (S)-2-haloacid dehalogenase, CALCIUM ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBL
 
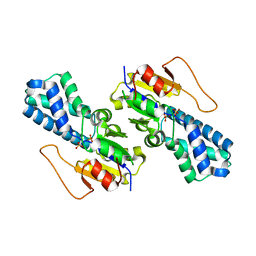 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] complexed with sulfate ions | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBO
 
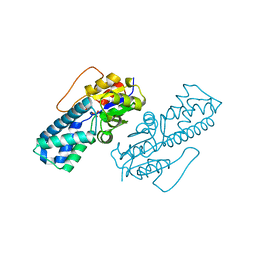 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant D18N complexed with sulfate ions | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBT
 
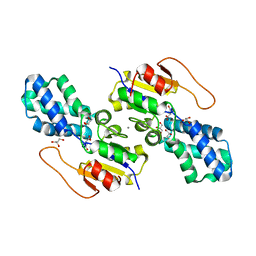 | | Crystal structure of cis-Epoxysuccinate Hydrolases KlCESH[L] mutant D48N complexed with L-TA | | Descriptor: | (S)-2-haloacid dehalogenase, CALCIUM ION, GLYCEROL, ... | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBP
 
 | |
8WBM
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant D193A complexed with sulfate ions | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBQ
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant E212Q complexed with L-TA. | | Descriptor: | Epoxide hydrolase, L(+)-TARTARIC ACID | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
4Q8K
 
 | | Crystal structure of polysaccharide lyase family 18 aly-SJ02 P-CATD | | Descriptor: | Alginase, CALCIUM ION, SULFATE ION | | Authors: | Dong, S, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2014-04-28 | | Release date: | 2014-09-17 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Molecular insight into the role of the N-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18.
J.Biol.Chem., 289, 2014
|
|
4Q8L
 
 | | Crystal structure of polysacchride lyase family 18 aly-SJ02 r-CATD | | Descriptor: | Alginase, CALCIUM ION | | Authors: | Dong, S, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2014-04-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Molecular insight into the role of the N-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18.
J.Biol.Chem., 289, 2014
|
|
7CCF
 
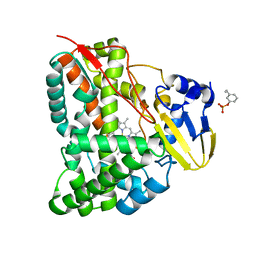 | | Mechanism insights on steroselective oxidation of phosphorylated ethylphenols with cytochrome P450 CreJ | | Descriptor: | (3-ethylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Du, L, Li, S.Y, Feng, Y.G. | | Deposit date: | 2020-06-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Selective Oxidation of Phosphorylated Ethylphenols by Cytochrome P450 Monooxygenase CreJ.
Appl.Environ.Microbiol., 87, 2021
|
|
8KC7
 
 | | Rpd3S histone deacetylase complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-06 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD7
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 167bp DNA | | Descriptor: | 167bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD4
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD2
 
 | | Rpd3S in complex with 187bp nucleosome | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD3
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD6
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD5
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
7CG5
 
 | |
7CG1
 
 | |
7CG8
 
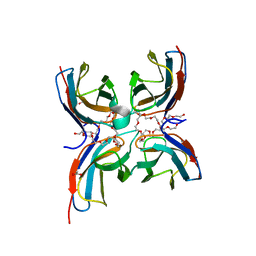 | | Structure of the sensor domain (short construct) of the anti-sigma factor RsgI4 in Pseudobacteroides cellulosolvens | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, ACETATE ION, Anti-sigma factor RsgI, ... | | Authors: | Dong, S, Feng, Y. | | Deposit date: | 2020-06-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the sensor domain of the anti-sigma factor RsgI4 in Pseudobacteroides cellulosolvens
To Be Published
|
|
5YJ7
 
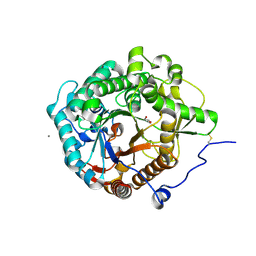 | | Structural insight into the beta-GH1 glucosidase BGLN1 from oleaginous microalgae Nannochloropsis | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside hydrolase | | Authors: | Dong, S, Liu, Y.J, Zhou, H.X, Xiao, Y, Xu, J, Cui, Q, Wang, X.Q, Feng, Y.G. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insight into a GH1 beta-glucosidase from the oleaginous microalga, Nannochloropsis oceanica.
Int.J.Biol.Macromol., 170, 2021
|
|
