1JU5
 
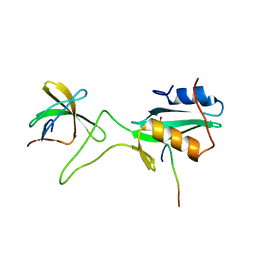 | | Ternary complex of an Crk SH2 domain, Crk-derived phophopeptide, and Abl SH3 domain by NMR spectroscopy | | Descriptor: | Abl, Crk | | Authors: | Donaldson, L.W, Pawson, T, Kay, L.E, Forman-Kay, J.D. | | Deposit date: | 2001-08-23 | | Release date: | 2002-11-06 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Structure of a regulatory complex involving the Abl SH3 domain, the Crk
SH2 domain, and a Crk-derived phosphopeptide
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2RNJ
 
 | | NMR Structure of The S. Aureus VraR DNA Binding Domain | | Descriptor: | Response regulator protein vraR | | Authors: | Donaldson, L.W. | | Deposit date: | 2008-01-09 | | Release date: | 2008-01-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of the Staphylococcus aureus Response Regulator VraR DNA Binding Domain Reveals a Dynamic Relationship between It and Its Associated Receiver Domain
Biochemistry, 47, 2008
|
|
1OVX
 
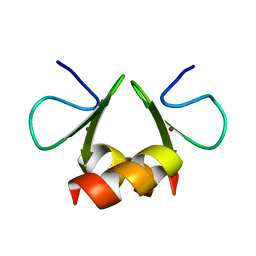 | | NMR structure of the E. coli ClpX chaperone zinc binding domain dimer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | Authors: | Donaldson, L.W, Kwan, J, Wojtyra, U, Houry, W.A. | | Deposit date: | 2003-03-27 | | Release date: | 2003-12-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dimeric zinc binding domain of the chaperone ClpX.
J.Biol.Chem., 278, 2003
|
|
6UZJ
 
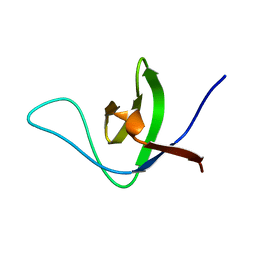 | | NMR structure of the HACS1 SH3 domain | | Descriptor: | SAM domain-containing protein SAMSN-1 | | Authors: | Donaldson, L.W. | | Deposit date: | 2019-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | HACS1 signaling adaptor protein recognizes a motif in the paired immunoglobulin receptor B cytoplasmic domain.
Commun Biol, 3, 2020
|
|
8DSB
 
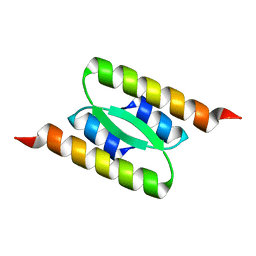 | | Lambda Bacteriophage Orf63 | | Descriptor: | Xis (Excision72) | | Authors: | Donaldson, L.W. | | Deposit date: | 2022-07-22 | | Release date: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Bad phages in good bacteria: Role of the mysterious orf63 of lambda and shiga toxin-converting phi24B bacteriophages.
Front Microbiol, 8, 2017
|
|
8DSX
 
 | |
5L1M
 
 | | CASKIN2 SAM domain tandem | | Descriptor: | Caskin-2 | | Authors: | Donaldson, L.W, Kwan, J.J, Saridakis, V. | | Deposit date: | 2016-07-29 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | A new mode of SAM domain mediated oligomerization observed in the CASKIN2 neuronal scaffolding protein.
Cell Commun. Signal, 14, 2016
|
|
2B6G
 
 | |
1OW5
 
 | |
2KIV
 
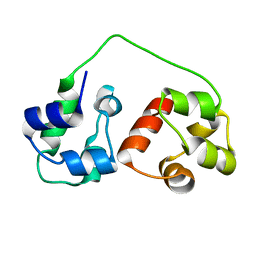 | | AIDA-1 SAM domain tandem | | Descriptor: | Ankyrin repeat and sterile alpha motif domain-containing protein 1B | | Authors: | Donaldson, L.W, Kurabi, A. | | Deposit date: | 2009-05-12 | | Release date: | 2009-08-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A nuclear localization signal at the SAM-SAM domain interface of AIDA-1 suggests a requirement for domain uncoupling prior to nuclear import.
J.Mol.Biol., 392, 2009
|
|
4IS7
 
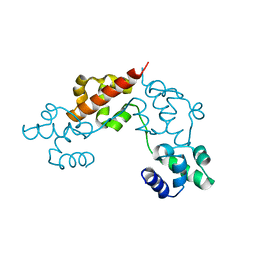 | |
1Z1Z
 
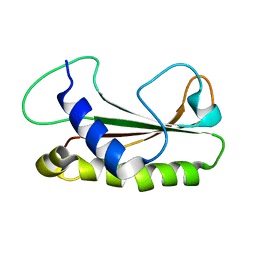 | |
2B7G
 
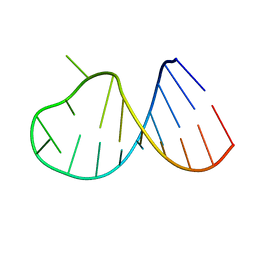 | |
2D3D
 
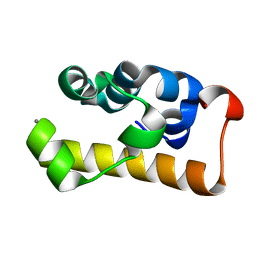 | | crystal structure of the RNA binding SAM domain of saccharomyces cerevisiae Vts1 | | Descriptor: | CALCIUM ION, Vts1 protein | | Authors: | Aviv, T, Amborski, A.N, Zhao, X.S, Kwan, J.J, Johnson, P.E, Sicheri, F, Donaldson, L.W. | | Deposit date: | 2005-09-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The NMR and X-ray Structures of the Saccharomyces cerevisiae Vts1 SAM Domain Define a Surface for the Recognition of RNA Hairpins
J.Mol.Biol., 356, 2006
|
|
1Z1V
 
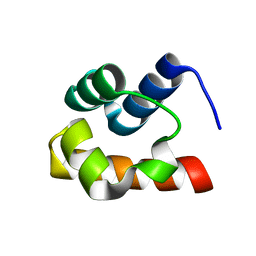 | | NMR structure of the Saccharomyces cerevisiae Ste50 SAM domain | | Descriptor: | STE50 protein | | Authors: | Kwan, J.J, Warner, N, Maini, J, Pawson, T, Donaldson, L.W. | | Deposit date: | 2005-03-06 | | Release date: | 2006-02-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Saccharomyces cerevisiae Ste50 binds the MAPKKK Ste11 through a head-to-tail SAM domain interaction.
J.Mol.Biol., 356, 2006
|
|
1R36
 
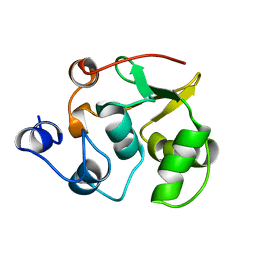 | | NMR-based structure of autoinhibited murine Ets-1 deltaN301 | | Descriptor: | C-ets-1 protein | | Authors: | Lee, G.M, Donaldson, L.W, Pufall, M.A, Kang, H.-S, Pot, I, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2003-09-30 | | Release date: | 2004-11-09 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The Structural and Dynamic Basis of Ets-1 DNA Binding Autoinhibition
J.Biol.Chem., 280, 2005
|
|
3FZ2
 
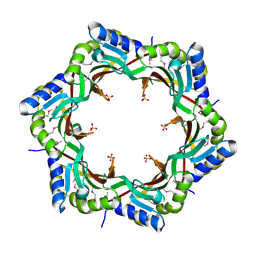 | | Crystal structure of the tail terminator protein from phage lambda (gpU-D74A) | | Descriptor: | Minor tail protein U, SULFATE ION | | Authors: | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | Deposit date: | 2009-01-23 | | Release date: | 2009-05-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
3FZB
 
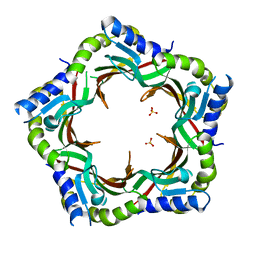 | | Crystal structure of the tail terminator protein from phage lambda (gpU-WT) | | Descriptor: | Minor tail protein U, SULFATE ION | | Authors: | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | Deposit date: | 2009-01-24 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
2L04
 
 | | The Solution Structure of the C-terminal Ig-like Domain of the Bacteriophage Lambda Tail Tube Protein | | Descriptor: | Major tail protein V | | Authors: | Pell, L.G, Gasmi-Seabrook, G.M.C, Donaldson, L.W, Howell, P, Davidson, A.R, Maxwell, K.L. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-22 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the C-Terminal Ig-like Domain of the Bacteriophage l Tail Tube Protein.
J.Mol.Biol., 403, 2010
|
|
2M38
 
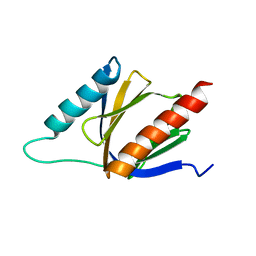 | | PTB domain of AIDA1 | | Descriptor: | Ankyrin repeat and sterile alpha motif domain-containing protein 1B | | Authors: | Donaldson, L. | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure and peptide binding of the PTB domain from the AIDA1 postsynaptic signaling scaffolding protein.
Plos One, 8, 2013
|
|
2M7B
 
 | |
2M7A
 
 | |
2K4Q
 
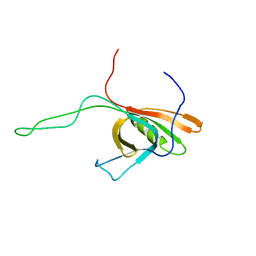 | | The Solution Structure of gpV, the Major Tail Protein from Bacteriophage Lambda | | Descriptor: | Major tail protein V | | Authors: | Pell, L.G, Kanelis, V, Howell, P, Davidson, A.R. | | Deposit date: | 2008-06-16 | | Release date: | 2009-02-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1BSH
 
 | |
1BSN
 
 | |
