8P64
 
 | | Co-crystal structure of PD-L1 with low molecular weight inhibitor | | Descriptor: | Programmed cell death 1 ligand 1, ~{N}-[[1-[(~{E})-2-(2-methyl-3-phenyl-phenyl)ethenyl]-1,2,3,4-tetrazol-5-yl]methyl]ethanamine | | Authors: | Plewka, J, Magiera-Mularz, K, van der Straat, R, Draijer, R, Surmiak, E, Butera, R, Land, L, Musielak, B, Domling, A. | | Deposit date: | 2023-05-25 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.312 Å) | | Cite: | 1,5-disubstituted Tetrazoles as PD-1/PD-L1 Antagonist
Rsc Med Chem, 2024
|
|
7BEA
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with inhibitor | | Descriptor: | 2-(aminomethyl)-6-[(2-methyl-3-phenyl-phenyl)methoxy]-~{N}-(2-phenylethyl)imidazo[1,2-a]pyridin-3-amine, Programmed cell death 1 ligand 1 | | Authors: | Magiera-Mularz, K, Butera, R, Wazynska, M, Holak, T, Domling, A. | | Deposit date: | 2020-12-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Imidazopyridines as PD-1/PD-L1 Antagonists.
Acs Med.Chem.Lett., 12, 2021
|
|
6ZRT
 
 | | Crystal structure of SARS CoV2 main protease in complex with inhibitor Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, DIMETHYL SULFOXIDE, Main Protease | | Authors: | Oerlemans, R, Wang, W, Lunev, S, Domling, A, Groves, M.R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Repurposing the HCV NS3-4A protease drug boceprevir as COVID-19 therapeutics.
Rsc Med Chem, 12, 2020
|
|
6ZRU
 
 | | Crystal structure of SARS CoV2 main protease in complex with inhibitor Boceprevir | | Descriptor: | DIMETHYL SULFOXIDE, Main Protease, boceprevir (bound form) | | Authors: | Oerlemans, R, Wang, W, Lunev, S, Domling, A, Groves, M.R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Repurposing the HCV NS3-4A protease drug boceprevir as COVID-19 therapeutics.
Rsc Med Chem, 12, 2020
|
|
5OAI
 
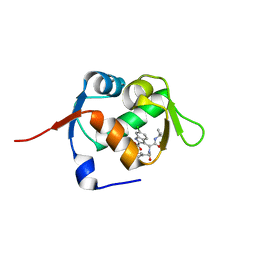 | | Structure of MDM2 with low molecular weight inhibitor | | Descriptor: | 3-[(1~{R})-2-(~{tert}-butylamino)-1-[methanoyl-[[3,4,5-tris(fluoranyl)phenyl]methyl]amino]-2-oxidanylidene-ethyl]-6-chloranyl-1~{H}-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Neochoritis, C.G, Grudnik, P, Dubin, G, Domling, A, Holak, T.A. | | Deposit date: | 2017-06-22 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A fluorinated indole-based MDM2 antagonist selectively inhibits the growth of p53wtosteosarcoma cells.
Febs J., 286, 2019
|
|
6R8G
 
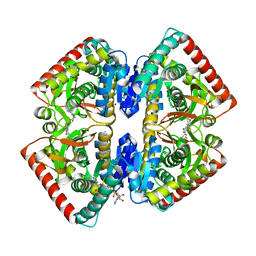 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with 4-(3,4-difluorophenyl)thiazol-2-amine | | Descriptor: | 4-[3,4-bis(fluoranyl)phenyl]-1,3-thiazol-2-amine, Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Fragment-Based Approach Identifies an Allosteric Pocket that Impacts Malate Dehydrogenase Activity
Commun Biol, 2021
|
|
7NT1
 
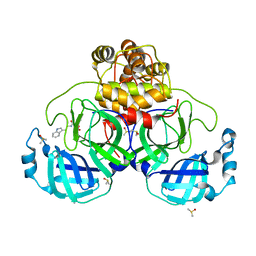 | | Crystal structure of SARS CoV2 main protease in complex with FSP007 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, [(2R)-1-[2-(1H-indol-3-yl)ethylamino]-1-oxidanylidene-butan-2-yl] prop-2-enoate | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NTV
 
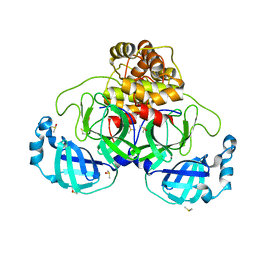 | | Crystal structure of SARS CoV2 main protease in complex with DN_EG_002 (modelled using PanDDA event map) | | Descriptor: | 2-acetamido-N-cyclopropyl-5-phenyl-thiophene-3-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NT2
 
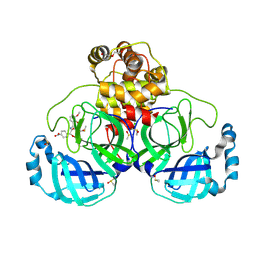 | | Crystal structure of SARS CoV2 main protease in complex with FSP006 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NUK
 
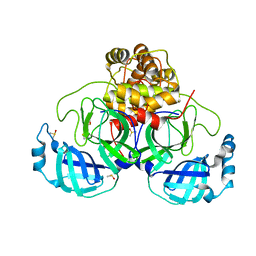 | | Crystal structure of SARS CoV2 main protease in complex with EG009 (modelled using PanDDA event map) | | Descriptor: | 2-[2-chloranylethanoyl(propyl)amino]-~{N}-(2-methoxyphenyl)ethanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NT3
 
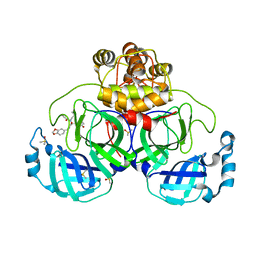 | | Crystal structure of SARS CoV2 main protease in complex with FSCU015 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(1~{S})-2-(1,3-benzodioxol-5-ylmethylamino)-1-(3-hydroxyphenyl)-2-oxidanylidene-ethyl]-~{N}-propyl-prop-2-enamide | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.325 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6T2F
 
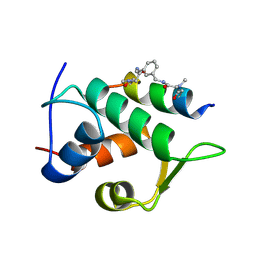 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | 2-(methylamino)-~{N}-[[3-[[2-(methylamino)ethanoylamino]methyl]phenyl]methyl]ethanamide, E3 ubiquitin-protein ligase Mdm2, MDM2 in complex with GAR300-Am | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6T2E
 
 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, Stapled peptide GAR300-Gm | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6T2D
 
 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | 2-(methylamino)-~{N}-[[4-[[2-(methylamino)ethanoylamino]methyl]phenyl]methyl]ethanamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6Y91
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with NADH | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity.
Commun Biol, 4, 2021
|
|
3TU1
 
 | | Exhaustive Fluorine Scanning towards Potent p53-MDM2 Antagonist | | Descriptor: | 2-(tert-butylamino)-1-(2-carboxy-6-chloro-1H-indol-3-yl)-1-[(3,4-difluorobenzyl)(formyl)amino]-2-oxoethylium, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Wolf, S, Huang, Y, Koes, D, Popowicz, G.M, Camacho, C.J, Holak, T.A, Doemling, A. | | Deposit date: | 2011-09-15 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Exhaustive Fluorine Scanning toward Potent p53-Mdm2 Antagonists.
Chemmedchem, 7, 2012
|
|
5J7F
 
 | | Structure of MDM2 with low molecular weight inhibitor with aliphatic linker. | | Descriptor: | 4-({6-[(6-chloro-3-{1-[(4-chlorophenyl)methyl]-4-(4-fluorophenyl)-1H-imidazol-5-yl}-1H-indole-2-carbonyl)oxy]hexyl}amino)-4-oxobutanoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Kubica, K, Guzik, K, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4,5-Trisubstituted Imidazole-Based p53-MDM2/MDMX Antagonists with Aliphatic Linkers for Conjugation with Biological Carriers.
J. Med. Chem., 60, 2017
|
|
5J7G
 
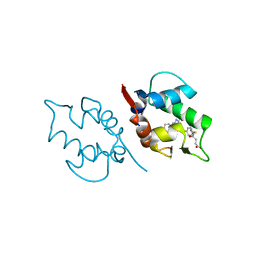 | | Structure of MDM2 with low molecular weight inhibitor with aliphatic linker. | | Descriptor: | 4-({6-[(6-chloro-3-{1-[(4-chlorophenyl)methyl]-4-(4-fluorophenyl)-1H-imidazol-5-yl}-1H-indole-2-carbonyl)oxy]hexyl}amino)-4-oxobutanoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Kubica, K, Guzik, K, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,4,5-Trisubstituted Imidazole-Based p53-MDM2/MDMX Antagonists with Aliphatic Linkers for Conjugation with Biological Carriers.
J. Med. Chem., 60, 2017
|
|
3LBL
 
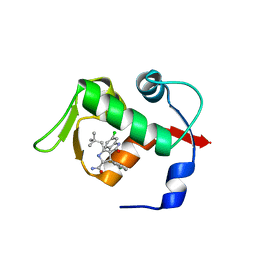 | | Structure of human MDM2 protein in complex with Mi-63-analog | | Descriptor: | (2'R,3R,4'R,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(2-morpholin-4-ylethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
3LBK
 
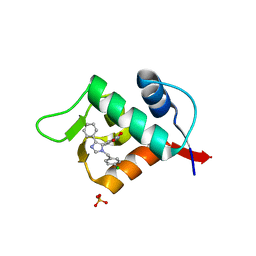 | | Structure of human MDM2 protein in complex with a small molecule inhibitor | | Descriptor: | 6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
3LBJ
 
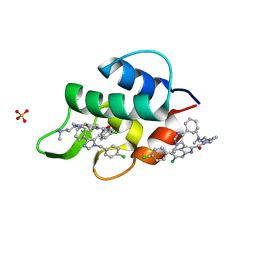 | | Structure of human MDMX protein in complex with a small molecule inhibitor | | Descriptor: | N-[(3S)-1-({6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indol-2-yl}carbonyl)pyrrolidin-3-yl]-N,N',N'-trimethylpropane-1,3-diamine, Protein Mdm4, SULFATE ION | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
4ZFI
 
 | | Structure of Mdm2 with low molecular weight inhibitor | | Descriptor: | (5S)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxy-1-methyl-1,5-dihydro-2H-pyrrol-2-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Zak, K.M, Twarda-Clapa, A, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2015-04-21 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
4ZGK
 
 | | Structure of Mdm2 with low molecular weight inhibitor. | | Descriptor: | (5R)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxyfuran-2(5H)-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Zak, K.M, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2015-04-23 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
4MDN
 
 | | Structure of a novel submicromolar MDM2 inhibitor | | Descriptor: | 3-{(1S)-2-(tert-butylamino)-1-[{4-[(4-chlorobenzyl)oxy]benzyl}(formyl)amino]-2-oxoethyl}-6-chloro-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Bista, M, Popowicz, G, Holak, T.A. | | Deposit date: | 2013-08-23 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
4MDQ
 
 | | Structure of a novel submicromolar MDM2 inhibitor | | Descriptor: | 3-[(1R)-2-(benzylamino)-1-{[(2S)-1-(hydroxyamino)-4-methyl-1-oxopentan-2-yl]amino}-2-oxoethyl]-6-chloro-N-hydroxy-1H-indole-2-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Bista, M, Popowicz, G, Holak, T.A. | | Deposit date: | 2013-08-23 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
