1DOM
 
 | |
1DON
 
 | |
1CRQ
 
 | | THE SOLUTION STRUCTURE AND DYNAMICS OF RAS P21. GDP DETERMINED BY HETERONUCLEAR THREE AND FOUR DIMENSIONAL NMR SPECTROSCOPY | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kraulis, P.J, Domaille, P.J, Campbell-Burk, S.L, Van Aken, T, Laue, E.D. | | Deposit date: | 1993-11-24 | | Release date: | 1994-07-31 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1CRR
 
 | | THE SOLUTION STRUCTURE AND DYNAMICS OF RAS P21. GDP DETERMINED BY HETERONUCLEAR THREE AND FOUR DIMENSIONAL NMR SPECTROSCOPY | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kraulis, P.J, Domaille, P.J, Campbell-Burk, S.L, Van Aken, T, Laue, E.D. | | Deposit date: | 1993-11-24 | | Release date: | 1994-07-31 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1CRP
 
 | | THE SOLUTION STRUCTURE AND DYNAMICS OF RAS P21. GDP DETERMINED BY HETERONUCLEAR THREE AND FOUR DIMENSIONAL NMR SPECTROSCOPY | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kraulis, P.J, Domaille, P.J, Campbell-Burk, S.L, Van Aken, T, Laue, E.D. | | Deposit date: | 1993-11-24 | | Release date: | 1994-07-31 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1K0X
 
 | |
1BLX
 
 | | P19INK4D/CDK6 COMPLEX | | Descriptor: | CALCIUM ION, CYCLIN-DEPENDENT KINASE 6, P19INK4D | | Authors: | Brotherton, D.H, Dhanaraj, V, Wick, S, Brizuela, L, Domaille, P.J, Volyanik, E, Xu, X, Parisini, E, Smith, B.O, Archer, S.J, Serrano, M, Brenner, S.L, Blundell, T.L, Laue, E.D. | | Deposit date: | 1998-07-21 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d.
Nature, 395, 1998
|
|
1CEY
 
 | | ASSIGNMENTS, SECONDARY STRUCTURE, GLOBAL FOLD, AND DYNAMICS OF CHEMOTAXIS Y PROTEIN USING THREE-AND FOUR-DIMENSIONAL HETERONUCLEAR (13C,15N) NMR SPECTROSCOPY | | Descriptor: | CHEY | | Authors: | Moy, F.J, Lowry, D.F, Matsumura, P, Dahlquist, F.W, Krywko, J.E, Domaille, P.J. | | Deposit date: | 1994-11-23 | | Release date: | 1995-02-07 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Assignments, secondary structure, global fold, and dynamics of chemotaxis Y protein using three- and four-dimensional heteronuclear (13C,15N) NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1AP7
 
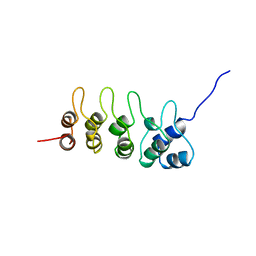 | | P19-INK4D FROM MOUSE, NMR, 20 STRUCTURES | | Descriptor: | P19-INK4D | | Authors: | Archer, S.J, Luh, F.Y, Domaille, P.J, Smith, B.O, Laue, E.D. | | Deposit date: | 1997-07-25 | | Release date: | 1998-09-16 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the cyclin-dependent kinase inhibitor p19Ink4d.
Nature, 389, 1997
|
|
1AP0
 
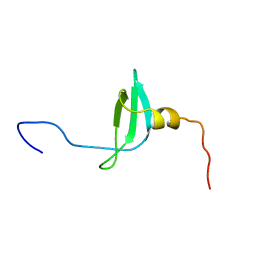 | | STRUCTURE OF THE CHROMATIN BINDING (CHROMO) DOMAIN FROM MOUSE MODIFIER PROTEIN 1, NMR, 26 STRUCTURES | | Descriptor: | MODIFIER PROTEIN 1 | | Authors: | Ball, L.J, Murzina, N.V, Broadhurst, R.W, Raine, A.R.C, Archer, S.J, Stott, F.J, Murzin, A.G, Singh, P.B, Domaille, P.J, Laue, E.D. | | Deposit date: | 1997-07-22 | | Release date: | 1998-07-22 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the chromatin binding (chromo) domain from mouse modifier protein 1.
EMBO J., 16, 1997
|
|
1BVE
 
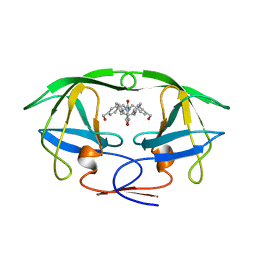 | | HIV-1 PROTEASE-DMP323 COMPLEX IN SOLUTION, NMR, 28 STRUCTURES | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Yamazaki, T, Hinck, A.P, Wang, Y.-X, Nicholson, L.K, Torchia, D.A, Wingfield, P, Stahl, S.J, Kaufman, J.D, Chang, C, Domaille, P.J, Lam, P.Y.S. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy.
Protein Sci., 5, 1996
|
|
1BVG
 
 | | HIV-1 PROTEASE-DMP323 COMPLEX IN SOLUTION, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Yamazaki, T, Hinck, A.P, Wang, Y.-X, Nicholson, L.K, Torchia, D.A, Wingfield, P, Stahl, S.J, Kaufman, J.D, Chang, C, Domaille, P.J, Lam, P.Y.S. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy.
Protein Sci., 5, 1996
|
|
1DOK
 
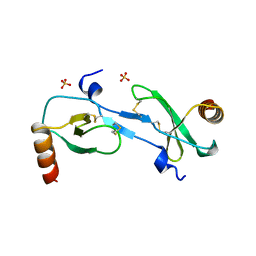 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, P-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1, SULFATE ION | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A, Domaille, P.J, Handel, T.M. | | Deposit date: | 1996-11-27 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
1DOL
 
 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, I-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1 | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A. | | Deposit date: | 1996-11-22 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
2KNX
 
 | |
2KNY
 
 | |
1GUW
 
 | | STRUCTURE OF THE CHROMODOMAIN FROM MOUSE HP1beta IN COMPLEX WITH THE LYSINE 9-METHYL HISTONE H3 N-TERMINAL PEPTIDE, NMR, 25 STRUCTURES | | Descriptor: | CHROMOBOX PROTEIN HOMOLOG 1, HISTONE H3.1 | | Authors: | Nielsen, P.R, Nietlispach, D, Mott, H.R, Callaghan, J.M, Bannister, A, Kouzarides, T, Murzin, A.G, Murzina, N.V, Laue, E.D. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-12 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | Structure of the Hp1 Chromodomain Bound to Histone H3 Methylated at Lysine 9
Nature, 416, 2002
|
|
