3GOV
 
 | | Crystal structure of the catalytic region of human MASP-1 | | Descriptor: | GLYCEROL, MASP-1 | | Authors: | Harmat, V, Dobo, J, Beinrohr, L, Sebestyen, E, Zavodszky, P, Gal, P. | | Deposit date: | 2009-03-20 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | MASP-1, a promiscuous complement protease: structure of its catalytic region reveals the basis of its broad specificity.
J.Immunol., 183, 2009
|
|
7ARX
 
 | | Crystal structure of the catalytic fragment of masp-1 in complex with SFMI1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Mannan-binding lectin serine protease 1, SFMI1 - Sunflower MASP1 inhibitor | | Authors: | Durvanger, Z, Harmat, V, Dobo, J, Megyeri, M. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Directed Evolution-Driven Increase of Structural Plasticity Is a Prerequisite for Binding the Complement Lectin Pathway Blocking MASP-Inhibitor Peptides.
Acs Chem.Biol., 17, 2022
|
|
4IGD
 
 | | Crystal structure of the zymogen catalytic region of Human MASP-1 | | Descriptor: | GLYCEROL, Mannan-binding lectin serine protease 1 | | Authors: | Harmat, V, Megyeri, M, Vegh, A, Dobo, J. | | Deposit date: | 2012-12-17 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quantitative characterization of the activation steps of mannan-binding lectin (MBL)-associated serine proteases (MASPs) points to the central role of MASP-1 in the initiation of the complement lectin pathway
J.Biol.Chem., 288, 2013
|
|
2D26
 
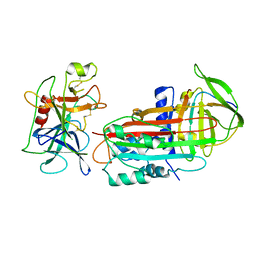 | |
2OAY
 
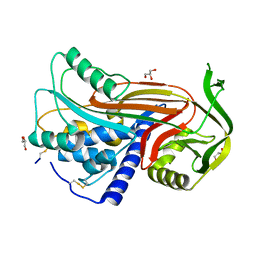 | | Crystal structure of latent human C1-inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Plasma protease C1 inhibitor | | Authors: | Harmat, V, Beinrohr, L, Gal, P, Dobo, J. | | Deposit date: | 2006-12-18 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | C1 inhibitor serpin domain structure reveals the likely mechanism of heparin potentiation and conformational disease
J.Biol.Chem., 282, 2007
|
|
3TVJ
 
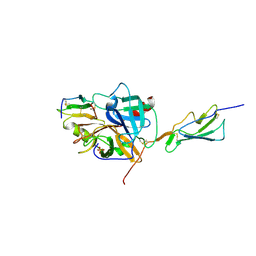 | | Catalytic fragment of MASP-2 in complex with its specific inhibitor developed by directed evolution on SGCI scaffold | | Descriptor: | Mannan-binding lectin serine protease 2 A chain, Mannan-binding lectin serine protease 2 B chain, Protease inhibitor SGPI-2, ... | | Authors: | Heja, D, Harmat, V, Dobo, J, Szasz, R, Kekesi, K.A, Zavodszky, P, Gal, P, Pal, G. | | Deposit date: | 2011-09-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Monospecific Inhibitors Show That Both Mannan-binding Lectin-associated Serine Protease-1 (MASP-1) and -2 Are Essential for Lectin Pathway Activation and Reveal Structural Plasticity of MASP-2.
J.Biol.Chem., 287, 2012
|
|
4DJZ
 
 | | Catalytic fragment of masp-1 in complex with its specific inhibitor developed by directed evolution on sgci scaffold | | Descriptor: | Mannan-binding lectin serine protease 1 heavy chain, Mannan-binding lectin serine protease 1 light chain, Protease inhibitor SGPI-2 | | Authors: | Heja, D, Harmat, V, Fodor, K, Wilmanns, M, Dobo, J, Kekesi, K.A, Zavodszky, P, Gal, P, Pal, G. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Monospecific Inhibitors Show That Both Mannan-binding Lectin-associated Serine Protease-1 (MASP-1) and -2 Are Essential for Lectin Pathway Activation and Reveal Structural Plasticity of MASP-2.
J.Biol.Chem., 287, 2012
|
|
7PQN
 
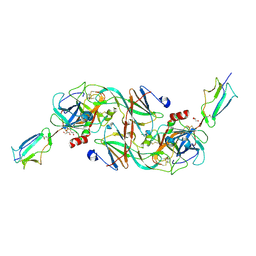 | | Catalytic fragment of MASP-2 in complex with ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 2 A chain, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.400015 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
7PQO
 
 | | Catalytic fragment of MASP-1 in complex with P1 site mutant ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 1, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
5JPM
 
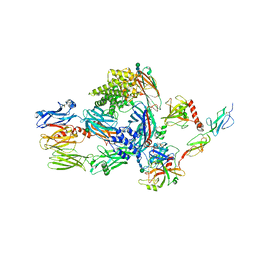 | | Structure of the complex of human complement C4 with MASP-2 rebuilt using iMDFF | | Descriptor: | Complement C4-A, Mannan-binding lectin serine protease 2, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Croll, T.I, Andersen, G.R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Re-evaluation of low-resolution crystal structures via interactive molecular-dynamics flexible fitting (iMDFF): a case study in complement C4.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5JPN
 
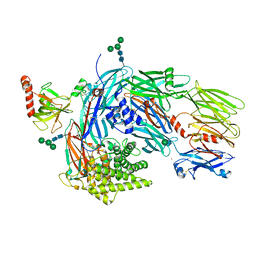 | | Structure of human complement C4 rebuilt using iMDFF | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C4-A, ... | | Authors: | Croll, T.I, Andersen, G.R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Re-evaluation of low-resolution crystal structures via interactive molecular-dynamics flexible fitting (iMDFF): a case study in complement C4.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
