1N5W
 
 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Oxidized form | | Descriptor: | CU(I)-S-MO(VI)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-07 | | Release date: | 2002-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N63
 
 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Carbon monoxide reduced state | | Descriptor: | CU(I)-S-MO(IV)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N61
 
 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Dithionite reduced state | | Descriptor: | CU(I)-S-MO(IV)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N62
 
 | | Crystal Structure of the Mo,Cu-CO Dehydrogenase (CODH), n-butylisocyanide-bound state | | Descriptor: | CU(I)-S-MO(IV)(=O)O-NBIC CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N60
 
 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Cyanide-inactivated Form | | Descriptor: | Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, Carbon monoxide dehydrogenase small chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1SU8
 
 | | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase | | Descriptor: | Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Dobbek, H, Svetlitchnyi, V, Liss, J, Meyer, O. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase
J.Am.Chem.Soc., 126, 2004
|
|
1SUF
 
 | | Carbon Monoxide Dehydrogenase from Carboxydothermus hydrogenoformans-Inactive state | | Descriptor: | Carbon Monoxide Dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Dobbek, H, Svetlitchnyi, V, Liss, J, Meyer, O. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase
J.Am.Chem.Soc., 126, 2004
|
|
1SU7
 
 | | Carbon Monoxide Dehydrogenase from Carboxydothermus hydrogenoformans- DTT reduced state | | Descriptor: | Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Dobbek, H, Svetlitchnyi, V, Liss, J, Meyer, O. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase
J.Am.Chem.Soc., 126, 2004
|
|
1SU6
 
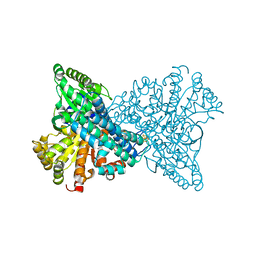 | | Carbon monoxide dehydrogenase from Carboxydothermus hydrogenoformans: CO reduced state | | Descriptor: | Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Dobbek, H, Svetlitchnyi, V, Liss, J, Meyer, O. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase
J.Am.Chem.Soc., 126, 2004
|
|
2OYY
 
 | | HTHP: a hexameric tyrosine-coordinated heme protein | | Descriptor: | Hexameric cytochrome, IODIDE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dobbek, H. | | Deposit date: | 2007-02-23 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | HTHP: A Novel Class of Hexameric, Tyrosine-coordinated Heme Proteins
J.Mol.Biol., 368, 2007
|
|
6T7J
 
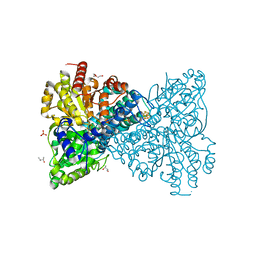 | | As-isolated Ni-free crystal structure of carbon monoxide dehydrogenase from Thermococcus sp. AM4 produced without CooC maturase | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Broken Fe4S4 cluster, CITRIC ACID, ... | | Authors: | Dobbek, H, Jeoung, J.-H. | | Deposit date: | 2019-10-22 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The two CO-dehydrogenases of Thermococcus sp. AM4.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
2H90
 
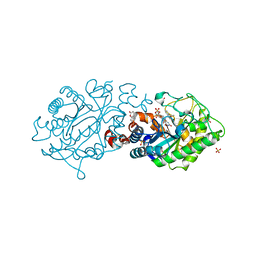 | | Xenobiotic reductase A in complex with coumarin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, COUMARIN, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobbek, H. | | Deposit date: | 2006-06-08 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Xenobiotic reductase A in the degradation of quinoline by Pseudomonas putida 86: physiological function, structure and mechanism of 8-hydroxycoumarin reduction.
J.Mol.Biol., 361, 2006
|
|
2H8Z
 
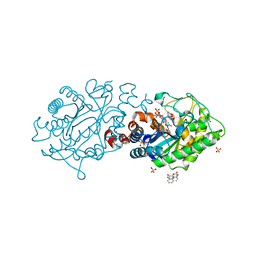 | | Xenobiotic Reductase A in complex with 8-Hydroxycoumarin | | Descriptor: | 8-HYDROXYCOUMARIN, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Dobbek, H. | | Deposit date: | 2006-06-08 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Xenobiotic reductase A in the degradation of quinoline by Pseudomonas putida 86: physiological function, structure and mechanism of 8-hydroxycoumarin reduction.
J.Mol.Biol., 361, 2006
|
|
2H8X
 
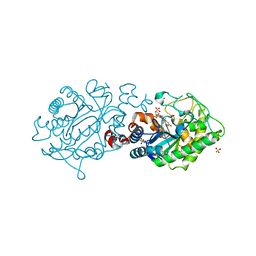 | | Xenobiotic Reductase A-oxidized | | Descriptor: | FLAVIN MONONUCLEOTIDE, SULFATE ION, Xenobiotic reductase A | | Authors: | Dobbek, H. | | Deposit date: | 2006-06-08 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Xenobiotic reductase A in the degradation of quinoline by Pseudomonas putida 86: physiological function, structure and mechanism of 8-hydroxycoumarin reduction.
J.Mol.Biol., 361, 2006
|
|
2H9A
 
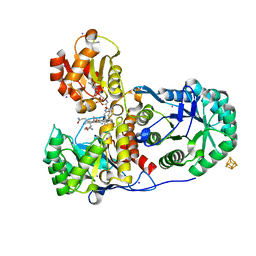 | | Corrinoid Iron-Sulfur Protein | | Descriptor: | CO dehydrogenase/acetyl-CoA synthase, iron-sulfur protein, COBALAMIN, ... | | Authors: | Dobbek, H. | | Deposit date: | 2006-06-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into methyltransfer reactions of a corrinoid iron-sulfur protein involved in acetyl-CoA synthesis.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2IHR
 
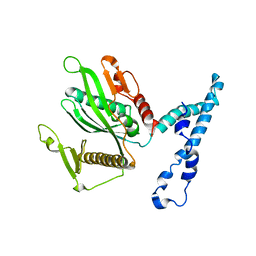 | | RF2 of Thermus thermophilus | | Descriptor: | Peptide chain release factor 2 | | Authors: | Dobbek, H, Voertler, C.S, Sprinzl, M. | | Deposit date: | 2006-09-27 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Release factors 2 from Escherichia coli and Thermus thermophilus: structural, spectroscopic and microcalorimetric studies.
Nucleic Acids Res., 35, 2007
|
|
3N14
 
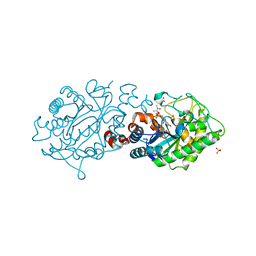 | | XenA - W358A | | Descriptor: | (R,R)-2,3-BUTANEDIOL, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Dobbek, H, Spiegelhauer, O. | | Deposit date: | 2010-05-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determinants of substrate binding and protonation in the flavoenzyme xenobiotic reductase A
J.Mol.Biol., 403, 2010
|
|
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5AA5
 
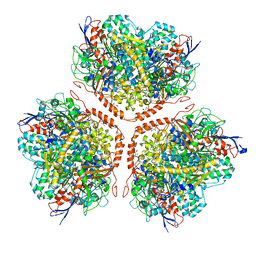 | | Actinobacterial-type NiFe-hydrogenase from Ralstonia eutropha H16 at 2.85 Angstrom resolution | | Descriptor: | IRON/SULFUR CLUSTER, MALONIC ACID, NIFE-HYDROGENASE LARGE SUBUNIT, ... | | Authors: | Schaefer, C, Bommer, M, Hennig, S, Jeoung, J.H, Dobbek, H, Lenz, O. | | Deposit date: | 2015-07-23 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structure of an Actinobacterial-Type [Nife]-Hydrogenase Reveals Insight Into O2-Tolerant H2 Oxidation.
Structure, 24, 2016
|
|
4PJ0
 
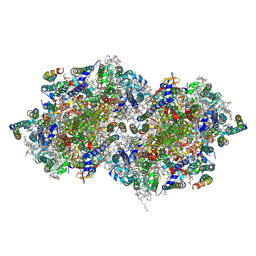 | | Structure of T.elongatus Photosystem II, rows of dimers crystal packing | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hellmich, J, Bommer, M, Burkhardt, A, Ibrahim, M, Kern, J, Meents, A, Mueh, F, Dobbek, H, Zouni, A. | | Deposit date: | 2014-05-10 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.437 Å) | | Cite: | Native-like Photosystem II Superstructure at 2.44 angstrom Resolution through Detergent Extraction from the Protein Crystal.
Structure, 22, 2014
|
|
1FFU
 
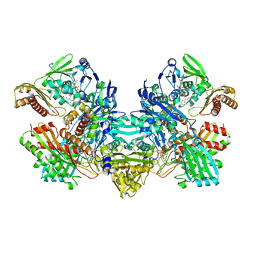 | | CARBON MONOXIDE DEHYDROGENASE FROM HYDROGENOPHAGA PSEUDOFLAVA WHICH LACKS THE MO-PYRANOPTERIN MOIETY OF THE MOLYBDENUM COFACTOR | | Descriptor: | CUTL, MOLYBDOPROTEIN OF CARBON MONOXIDE DEHYDROGENASE, CUTM, ... | | Authors: | Haenzelmann, P, Dobbek, H, Gremer, L, Huber, R, Meyer, O. | | Deposit date: | 2000-07-26 | | Release date: | 2000-09-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
J.Mol.Biol., 301, 2000
|
|
1FFV
 
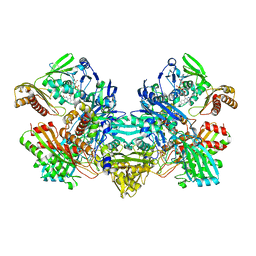 | | CARBON MONOXIDE DEHYDROGENASE FROM HYDROGENOPHAGA PSEUDOFLAVA | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), CUTL, MOLYBDOPROTEIN OF CARBON MONOXIDE DEHYDROGENASE, ... | | Authors: | Haenzelmann, P, Dobbek, H, Gremer, L, Huber, R, Meyer, O. | | Deposit date: | 2000-07-26 | | Release date: | 2000-09-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
J.Mol.Biol., 301, 2000
|
|
4UTH
 
 | | XenA - oxidized - Y183F variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase, SULFATE ION | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
8CJB
 
 | | A268M variant of the CODH/ACS complex of C. hydrogenoformans | | Descriptor: | ACETATE ION, CO-methylating acetyl-CoA synthase, Carbon monoxide dehydrogenase, ... | | Authors: | Ruickoldt, J, Jeoung, J, Lennartz, F, Dobbek, H. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A225L variant of the CODH/ACS complex of C. hydrogenoformans
To Be Published
|
|
8CJA
 
 | | A225L/F231A variant of the CODH/ACS complex of C. hydrogenoformans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CO-methylating acetyl-CoA synthase, ... | | Authors: | Ruickoldt, J, Jeoung, J, Lennartz, F, Dobbek, H. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A225L variant of the CODH/ACS complex of C. hydrogenoformans
To Be Published
|
|
