5OF1
 
 | | The structural versatility of TasA in B. subtilis biofilm formation | | Descriptor: | 2-HYDROXYBENZOIC ACID, Spore coat-associated protein N, ethane-1,2-diol | | Authors: | Roske, Y, Diehl, A, Ball, L, Chowdhury, A, Hiller, M, Moliere, N, Kramer, R, Nagaraj, M, Stoeppler, D, Worth, C.L, Schlegel, B, Leidert, M, Cremer, N, Eisenmenger, F, Lopez, D, Schmieder, P, Heinemann, U, Turgay, K, Akbey, U, Oschkinat, H. | | Deposit date: | 2017-07-10 | | Release date: | 2018-03-21 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural changes of TasA in biofilm formation ofBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OF2
 
 | | The structural versatility of TasA in B. subtilis biofilm formation | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Spore coat-associated protein N | | Authors: | Roske, Y, Diehl, A, Ball, L, Chowdhury, A, Hiller, M, Moliere, N, Kramer, R, Nagaraj, M, Stoeppler, D, Worth, C.L, Schlegel, B, Leidert, M, Cremer, N, Eisenmenger, F, Lopez, D, Schmieder, P, Heinemann, U, Turgay, K, Akbey, U, Oschkinat, H. | | Deposit date: | 2017-07-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural changes of TasA in biofilm formation ofBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1FLG
 
 | | CRYSTAL STRUCTURE OF THE QUINOPROTEIN ETHANOL DEHYDROGENASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | CALCIUM ION, PROTEIN (QUINOPROTEIN ETHANOL DEHYDROGENASE), PYRROLOQUINOLINE QUINONE | | Authors: | Keitel, T, Diehl, A, Knaute, T, Stezowski, J.J, Hohne, W, Gorisch, H. | | Deposit date: | 2000-08-14 | | Release date: | 2000-08-30 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the quinoprotein ethanol dehydrogenase from Pseudomonas aeruginosa: basis of substrate specificity.
J.Mol.Biol., 297, 2000
|
|
6QAY
 
 | |
1M8M
 
 | | SOLID-STATE MAS NMR STRUCTURE OF THE A-SPECTRIN SH3 DOMAIN | | Descriptor: | SPECTRIN ALPHA CHAIN, BRAIN | | Authors: | Castellani, F, Van Rossum, B, Diehl, A, Schubert, M, Rehbein, K, Oschkinat, H. | | Deposit date: | 2002-07-25 | | Release date: | 2002-11-20 | | Last modified: | 2022-12-21 | | Method: | SOLID-STATE NMR | | Cite: | Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy
Nature, 420, 2002
|
|
1M7K
 
 | | Solution Structure of the SODD BAG Domain | | Descriptor: | Silencer of Death Domains | | Authors: | Brockmann, C, Leitner, D, Labudde, D, Diehl, A, Sievert, V, Buessow, K, Oschkinat, H. | | Deposit date: | 2002-07-22 | | Release date: | 2002-08-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the SODD BAG domain reveals additional electrostatic interactions in the HSP70 complexes of SODD subfamily BAG domains
Febs Lett., 558, 2004
|
|
1S3A
 
 | | NMR Solution Structure of Subunit B8 from Human NADH-Ubiquinone Oxidoreductase Complex I (CI-B8) | | Descriptor: | NADH-ubiquinone oxidoreductase B8 subunit | | Authors: | Brockmann, C, Diehl, A, Rehbein, K, Kuhne, R, Oschkinat, H. | | Deposit date: | 2004-01-13 | | Release date: | 2005-01-25 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The oxidized subunit B8 from human complex I adopts a thioredoxin fold.
Structure, 12, 2004
|
|
1NEG
 
 | | Crystal Structure Analysis of N-and C-terminal labeled SH3-domain of alpha-Chicken Spectrin | | Descriptor: | AZIDE ION, Spectrin alpha chain, brain | | Authors: | Mueller, U, Buessow, K, Diehl, A, Niesen, F.H, Nyarsik, L, Heinemann, U. | | Deposit date: | 2002-12-11 | | Release date: | 2003-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid purification and crystal structure analysis of a small protein carrying two terminal affinity tags
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
1PQS
 
 | | Solution structure of the C-terminal OPCA domain of yCdc24p | | Descriptor: | Cell division control protein 24 | | Authors: | Leitner, D, Wahl, M, Labudde, D, Diehl, A, Schmieder, P, Pires, J.R, Fossi, M, Leidert, M, Krause, G, Oschkinat, H. | | Deposit date: | 2003-06-19 | | Release date: | 2003-07-01 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an N-terminally truncated version of the yeast CDC24p PB1 domain shows a different beta-sheet topology.
Febs Lett., 579, 2005
|
|
5KGQ
 
 | | NMR structure and dynamics of Q4DY78, a conserved kinetoplasid-specific protein from Trypanosoma cruzi | | Descriptor: | Uncharacterized protein | | Authors: | D'Andrea, E.D, Retel, J.S, Diehl, A, Schmieder, P, Oschkinat, H, Pires, J.R. | | Deposit date: | 2016-06-13 | | Release date: | 2017-07-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of Q4DY78, a conserved kinetoplasid-specific protein from Trypanosoma cruzi
To Be Published
|
|
1U06
 
 | | crystal structure of chicken alpha-spectrin SH3 domain | | Descriptor: | AZIDE ION, Spectrin alpha chain, brain | | Authors: | Chevelkov, V, Faelber, K, Diehl, A, Heinemann, U, Oschkinat, H, Reif, B. | | Deposit date: | 2004-07-13 | | Release date: | 2005-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Detection of dynamic water molecules in a microcrystalline sample of the SH3 domain of alpha-spectrin by MAS solid-state NMR.
J.Biomol.Nmr, 31, 2005
|
|
3O5N
 
 | | Tetrahydroquinoline carboxylates are potent inhibitors of the Shank PDZ domain, a putative target in autism disorders | | Descriptor: | (3aS,4R,9bR)-9-nitro-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-4,6-dicarboxylic acid, SH3 and multiple ankyrin repeat domains protein 3 | | Authors: | Saupe, J, Roske, Y, Schillinger, C, Kamdem, N, Radetzki, S, Diehl, A, Oschkinat, H, Krause, G, Heinemann, U, Rademann, J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery, structure-activity relationship studies, and crystal structure of nonpeptide inhibitors bound to the shank3 PDZ domain.
Chemmedchem, 6, 2011
|
|
2JRZ
 
 | | Solution structure of the Bright/ARID domain from the human JARID1C protein. | | Descriptor: | Histone demethylase JARID1C | | Authors: | Koehler, C, Bishop, S, Dowler, E.F, Diehl, A, Schmieder, P, Leidert, M, Sundstrom, M, Arrowsmith, C.H, Wiegelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Backbone and sidechain 1H, 13C and 15N resonance assignments of the Bright/ARID domain from the human JARID1C (SMCX) protein.
Biomol.Nmr Assign., 2, 2008
|
|
2I9S
 
 | | The solution structure of the core of mesoderm development (MESD). | | Descriptor: | Mesoderm development candidate 2 | | Authors: | Koehler, C, Andersen, O, Diehl, A, Schmieder, P, Krause, G, Oschkinat, H. | | Deposit date: | 2006-09-06 | | Release date: | 2007-05-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the core of mesoderm development (MESD), a chaperone for members of the LDLR-family
J.STRUCT.FUNCT.GENOM., 7, 2006
|
|
2JTF
 
 | | Solution Structure of the PHF20L1 MBT domain | | Descriptor: | PHD finger protein 20-like 1 | | Authors: | Brockmann, C, Iberg, A.N, Rehbein, K, Diehl, A, Bedford, M.T, Oschkinat, H. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of Histone H4K20 Methyllysine Recognition by the MBT Domain of PHF20L1
To be Published
|
|
2NUZ
 
 | | crystal structure of alpha spectrin SH3 domain measured at room temperature | | Descriptor: | Spectrin alpha chain, brain | | Authors: | Agarwal, V, Faelber, K, Hologne, M, Chevelkov, V, Oschkinat, H, Diehl, A, Reif, B. | | Deposit date: | 2006-11-10 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of MAS solid-state NMR and X-ray crystallographic data in the analysis of protein dynamics in the solid state
To be Published
|
|
2RQK
 
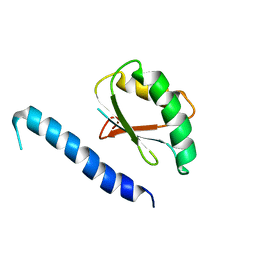 | | NMR Solution Structure of Mesoderm Development (MESD) - closed conformation | | Descriptor: | Mesoderm development candidate 2 | | Authors: | Koehler, C, Lighthouse, J.K, Werther, T, Andersen, O.M, Diehl, A, Schmieder, P, Holdener, B.C, Oschkinat, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The Structure of MESD45-184 Brings Light into the Mechanism of LDLR Family Folding
Structure, 19, 2011
|
|
2RQM
 
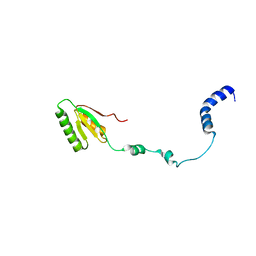 | | NMR Solution Structure of Mesoderm Development (MESD) - open conformation | | Descriptor: | Mesoderm development candidate 2 | | Authors: | Koehler, C, Lighthouse, J.K, Werther, T, Andersen, O.M, Diehl, A, Schmieder, P, Holdener, B.C, Oschkinat, H. | | Deposit date: | 2009-08-14 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The Structure of MESD45-184 Brings Light into the Mechanism of LDLR Family Folding
Structure, 19, 2011
|
|
2JM5
 
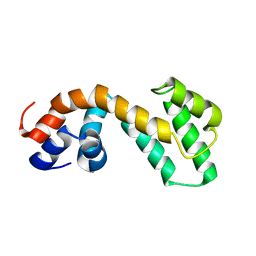 | | Solution Structure of the RGS domain from human RGS18 | | Descriptor: | Regulator of G-protein signaling 18 | | Authors: | Higman, V.A, Leidert, M, Bray, J, Elkins, J, Soundararajan, M, Doyle, D.A, Gileadi, C, Phillips, C, Schoch, G, Yang, X, Brockmann, C, Schmieder, P, Diehl, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-11 | | Release date: | 2006-10-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2OWI
 
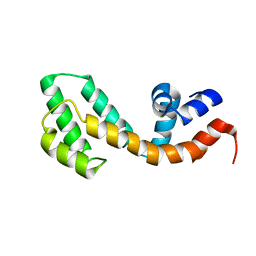 | | Solution structure of the RGS domain from human RGS18 | | Descriptor: | Regulator of G-protein signaling 18 | | Authors: | Higman, V.A, Leidert, M, Bray, J, Elkins, J, Soundararajan, M, Doyle, D.A, Gileadi, C, Phillips, C, Schoch, G, Yang, X, Brockmann, C, Schmieder, P, Diehl, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-16 | | Release date: | 2007-02-27 | | Last modified: | 2012-08-15 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6HQC
 
 | |
1SS6
 
 | | Solution structure of SEP domain from human p47 | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Soukenik, M, Leidert, M, Sievert, V, Buessow, K, Leitner, D, Labudde, D, Ball, L.J, Oschkinat, H. | | Deposit date: | 2004-03-23 | | Release date: | 2004-11-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The SEP domain of p47 acts as a reversible competitive inhibitor of cathepsin L
FEBS Lett., 576, 2004
|
|
8AIF
 
 | |
6ZC8
 
 | |
6ZBQ
 
 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 5-fluoranyl-2-(5,6,7,8-tetrahydronaphthalen-2-ylsulfonylamino)benzoic acid, Dishevelled, dsh homolog 3 (Drosophila), ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
