1S6L
 
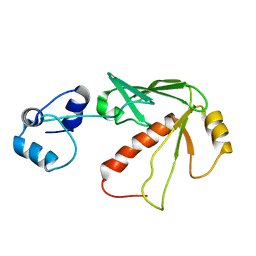 | | Solution structure of MerB, the Organomercurial Lyase involved in the bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase | | Authors: | Di Lello, P, Benison, G.C, Valafar, H, Pitts, K.E, Summers, A.O, Legault, P, Omichinski, J.G. | | Deposit date: | 2004-01-25 | | Release date: | 2005-04-19 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies reveal a novel protein fold for MerB, the organomercurial lyase involved in the bacterial mercury resistance system.
Biochemistry, 43, 2004
|
|
1Y5O
 
 | | NMR structure of the amino-terminal domain from the Tfb1 subunit of yeast TFIIH | | Descriptor: | RNA polymerase II transcription factor B 73 kDa subunit | | Authors: | Di Lello, P, Nguyen, B.D, Jones, T.N, Potempa, K, Kobor, M.S, Legault, P, Omichinski, J.G. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-17 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Amino-Terminal Domain from the Tfb1 Subunit of TFIIH and
Characterization of Its Phosphoinositide and VP16 Binding Sites
Biochemistry, 44, 2005
|
|
2GS0
 
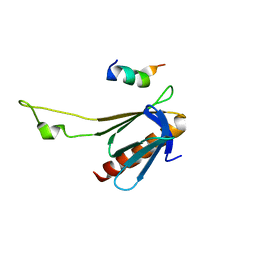 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the activation domain of p53 | | Descriptor: | Cellular tumor antigen p53, RNA polymerase II transcription factor B subunit 1 | | Authors: | Di Lello, P, Jones, T.N, Nguyen, B.D, Legault, P, Omichinski, J.G. | | Deposit date: | 2006-04-25 | | Release date: | 2006-10-31 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53.
Mol.Cell, 22, 2006
|
|
2JTX
 
 | |
1NJQ
 
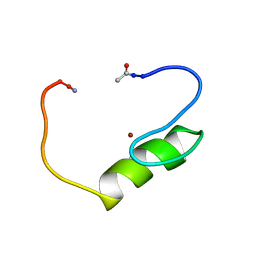 | | NMR structure of the single QALGGH zinc finger domain from Arabidopsis thaliana SUPERMAN protein | | Descriptor: | ZINC ION, superman protein | | Authors: | Isernia, C, Bucci, E, Leone, M, Zaccaro, L, Di Lello, P, Digilio, G, Esposito, S, Saviano, M, Di Blasio, B, Pedone, C, Pedone, P.V, Fattorusso, R. | | Deposit date: | 2003-01-02 | | Release date: | 2003-03-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Single QALGGH Zinc Finger Domain from the Arabidopsis thaliana SUPERMAN Protein.
Chembiochem, 4, 2003
|
|
1RQM
 
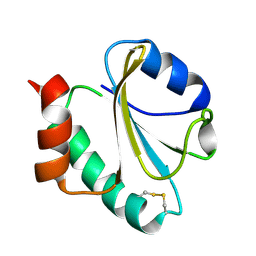 | | SOLUTION STRUCTURE OF THE K18G/R82E ALICYCLOBACILLUS ACIDOCALDARIUS THIOREDOXIN MUTANT | | Descriptor: | Thioredoxin | | Authors: | Leone, M, Di Lello, P, Ohlenschlager, O, Pedone, E.M, Bartolucci, S, Rossi, M, Di Blasio, B, Pedone, C, Saviano, M, Isernia, C, Fattorusso, R. | | Deposit date: | 2003-12-05 | | Release date: | 2004-06-22 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the K18G/R82E Alicyclobacillus acidocaldarius Thioredoxin Mutant: A Molecular Analysis of Its Reduced Thermal Stability.
Biochemistry, 43, 2004
|
|
2KDT
 
 | | PC1/3 DCSG sorting domain structure in DPC | | Descriptor: | Neuroendocrine convertase 1 | | Authors: | Dikeakos, J.D, Di Lello, P, Lacombe, M.J, Ghirlando, R, Legault, P, Reudelhuber, T.L, Omichinski, J.G. | | Deposit date: | 2009-01-19 | | Release date: | 2009-04-07 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Functional and structural characterization of a dense core secretory granule sorting domain from the PC1/3 protease.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2K7L
 
 | | NMR structure of a complex formed by the C-terminal domain of human RAP74 and a phosphorylated peptide from the central domain of the FCP1 | | Descriptor: | General transcription factor IIF subunit 1, centFCP1-T584PO4 peptide | | Authors: | Yang, A, Abbott, K.L, Desjardins, A, Di Lello, P, Omichinski, J.G, Legault, P. | | Deposit date: | 2008-08-13 | | Release date: | 2009-06-02 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a complex formed by the carboxyl-terminal domain of human RAP74 and a phosphorylated peptide from the central domain of the FCP1 phosphatase
Biochemistry, 48, 2009
|
|
2KE3
 
 | | PC1/3 DCSG sorting domain in CHAPS | | Descriptor: | Neuroendocrine convertase 1 | | Authors: | Dikeakos, J.D, Di Lello, P, Lacombe, M.J, Ghirlando, R, Legault, P, Reudelhuber, T.L, Omichinski, J.G. | | Deposit date: | 2009-01-22 | | Release date: | 2009-04-14 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Functional and structural characterization of a dense core secretory granule sorting domain from the PC1/3 protease
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2K2U
 
 | | NMR Structure of the complex between Tfb1 subunit of TFIIH and the activation domain of VP16 | | Descriptor: | Alpha trans-inducing protein, RNA polymerase II transcription factor B subunit 1 | | Authors: | Langlois, C, Mas, C, Di Lello, P, Miller Jenkins, P.M, Legault, J, Omichinski, J.G. | | Deposit date: | 2008-04-11 | | Release date: | 2008-08-12 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Complex between the Tfb1 Subunit of TFIIH and the Activation Domain of VP16: Structural Similarities between VP16 and p53.
J.Am.Chem.Soc., 130, 2008
|
|
3F0P
 
 | | Crystal structure of the mercury-bound form of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-25 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F0O
 
 | | Crystal structure of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, BROMIDE ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-25 | | Release date: | 2008-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F2H
 
 | | Crystal structure of the mercury-bound form of MerB mutant C160S, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, MERCURY (II) ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F2G
 
 | | Crystal structure of MerB mutant C160S, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F2F
 
 | | Crystal structure of the mercury-bound form of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
5IO9
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IP4
 
 | | X-RAY STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin, XA4551 NANOBODY AGAINST C-DCX | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IN7
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-23 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IOI
 
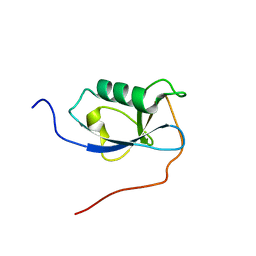 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
2L2I
 
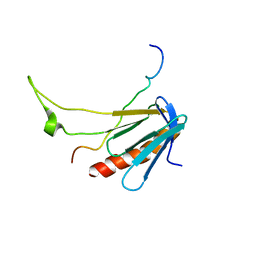 | |
5WHC
 
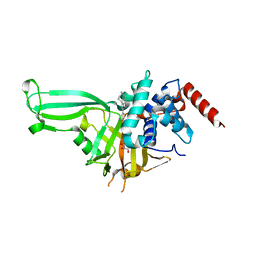 | | USP7 in complex with Cpd2 (4-(3-(1-methylpiperidin-4-yl)-1,2,4-oxadiazol-5-yl)phenol) | | Descriptor: | 4-[3-(1-methylpiperidin-4-yl)-1,2,4-oxadiazol-5-yl]phenol, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2017-07-16 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | Discovery of Small-Molecule Inhibitors of Ubiquitin Specific Protease 7 (USP7) Using Integrated NMR and in Silico Techniques.
J. Med. Chem., 60, 2017
|
|
4NMX
 
 | |
7TYU
 
 | | TEAD2 bound to Compound 2 | | Descriptor: | (3R)-1-[(8S)-5-(4-cyclohexylphenyl)-7-oxo-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonyl]pyrrolidine-3-carbonitrile, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
7TYP
 
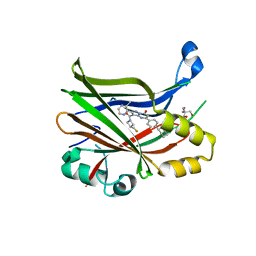 | | TEAD2 bound to GNE-7883 | | Descriptor: | (8S)-5-(4-cyclohexylphenyl)-3-[3-(fluoromethyl)azetidine-1-carbonyl]-2-(3-methylpyrazin-2-yl)pyrazolo[1,5-a]pyrimidin-7(4H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
7TYQ
 
 | | TEAD2 bound to Compound 1 | | Descriptor: | Transcriptional enhancer factor TEF-4, ethyl (8S)-7-oxo-5-[4-(trifluoromethyl)phenyl]-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carboxylate | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
