4DPM
 
 | | Structure of malonyl-coenzyme A reductase from crenarchaeota in complex with CoA | | Descriptor: | COENZYME A, MAGNESIUM ION, Malonyl-CoA/succinyl-CoA reductase | | Authors: | Demmer, U, Warkentin, E, Srivastava, A, Kockelkorn, D, Fuchs, G, Ermler, U. | | Deposit date: | 2012-02-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for a Bispecific NADP+ and CoA Binding Site in an Archaeal Malonyl-Coenzyme A Reductase.
J.Biol.Chem., 288, 2013
|
|
4DPK
 
 | | Structure of malonyl-coenzyme A reductase from crenarchaeota | | Descriptor: | Malonyl-CoA/succinyl-CoA reductase, PHOSPHATE ION | | Authors: | Demmer, U, Warkentin, E, Srivastava, A, Kockelkorn, D, Fuchs, G, Ermler, U. | | Deposit date: | 2012-02-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for a Bispecific NADP+ and CoA Binding Site in an Archaeal Malonyl-Coenzyme A Reductase.
J.Biol.Chem., 288, 2013
|
|
4DPL
 
 | | Structure of malonyl-coenzyme A reductase from crenarchaeota in complex with NadP | | Descriptor: | Malonyl-CoA/succinyl-CoA reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UNKNOWN LIGAND | | Authors: | Demmer, U, Warkentin, E, Srivastava, A, Kockelkorn, D, Fuchs, G, Ermler, U. | | Deposit date: | 2012-02-13 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for a Bispecific NADP+ and CoA Binding Site in an Archaeal Malonyl-Coenzyme A Reductase.
J.Biol.Chem., 288, 2013
|
|
1Z47
 
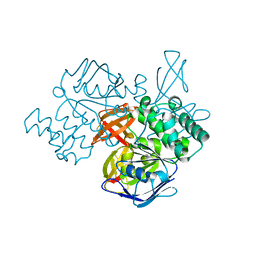 | | Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius | | Descriptor: | CHLORIDE ION, putative ABC-transporter ATP-binding protein | | Authors: | Scheffel, F, Demmer, U, Warkentin, E, Huelsmann, A, Schneider, E, Ermler, U. | | Deposit date: | 2005-03-15 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius
Febs Lett., 579, 2005
|
|
4YRY
 
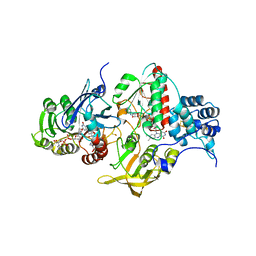 | | Insights into flavin-based electron bifurcation via the NADH-dependent reduced ferredoxin-NADP oxidoreductase structure | | Descriptor: | Dihydroorotate dehydrogenase B (NAD(+)), electron transfer subunit homolog, Dihydropyrimidine dehydrogenase subunit A, ... | | Authors: | Ermler, U, Thauer, R.K, Demmer, J.K, Huang, H, Wang, S, Demmer, U. | | Deposit date: | 2015-03-16 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into Flavin-based Electron Bifurcation via the NADH-dependent Reduced Ferredoxin:NADP Oxidoreductase Structure.
J.Biol.Chem., 290, 2015
|
|
4YLF
 
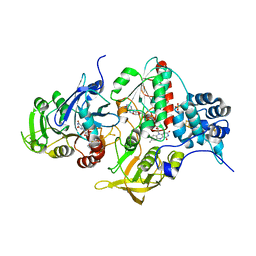 | | Insights into flavin-based electron bifurcation via the NADH-dependent reduced ferredoxin-NADP oxidoreductase structure | | Descriptor: | Dihydroorotate dehydrogenase B (NAD(+)), electron transfer subunit homolog, Dihydropyrimidine dehydrogenase subunit A, ... | | Authors: | Ermler, U, Thauer, R.K, Demmer, J.K, Huang, H, Wang, S, Demmer, U. | | Deposit date: | 2015-03-05 | | Release date: | 2015-07-08 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Insights into Flavin-based Electron Bifurcation via the NADH-dependent Reduced Ferredoxin:NADP Oxidoreductase Structure.
J.Biol.Chem., 290, 2015
|
|
4KPU
 
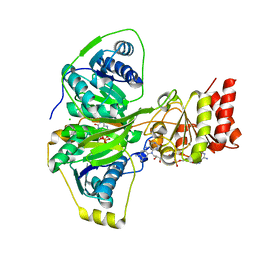 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | CHLORIDE ION, Electron transfer flavoprotein alpha subunit, Electron transfer flavoprotein alpha/beta-subunit, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-05-14 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
4L2I
 
 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | CHLORIDE ION, Electron transfer flavoprotein alpha subunit, Electron transfer flavoprotein alpha/beta-subunit, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-06-04 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
4L1F
 
 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | 1,3-PROPANDIOL, Acyl-CoA dehydrogenase domain protein, COENZYME A PERSULFIDE, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-06-03 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
2Y0F
 
 | | STRUCTURE OF GCPE (IspG) FROM THERMUS THERMOPHILUS HB27 | | Descriptor: | 4-HYDROXY-3-METHYLBUT-2-EN-1-YL DIPHOSPHATE SYNTHASE, IRON/SULFUR CLUSTER | | Authors: | Rekittke, I, Nonaka, T, Wiesner, J, Demmer, U, Warkentin, E, Jomaa, H, Ermler, U. | | Deposit date: | 2010-12-02 | | Release date: | 2011-01-26 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the E-1-Hydroxy-2-Methyl-But-2-Enyl-4-Diphosphate Synthase (Gcpe) from Thermus Thermophilus.
FEBS Lett., 585, 2011
|
|
7PXP
 
 | | Benzoylsuccinyl-CoA thiolase | | Descriptor: | Benzoylsuccinyl-CoA thiolase subunit, ZINC ION | | Authors: | Ermler, U, Heider, H, Weidenweber, S, Demmer, U. | | Deposit date: | 2021-10-08 | | Release date: | 2022-04-06 | | Last modified: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Finis tolueni: a new type of thiolase with an integrated Zn-finger subunit catalyzes the final step of anaerobic toluene metabolism.
Febs J., 289, 2022
|
|
6FAH
 
 | | Molecular basis of the flavin-based electron-bifurcating caffeyl-CoA reductase reaction | | Descriptor: | Caffeyl-CoA reductase-Etf complex subunit CarC, Caffeyl-CoA reductase-Etf complex subunit CarD, Caffeyl-CoA reductase-Etf complex subunit CarE, ... | | Authors: | Demmer, J.K, Bertsch, J, Oeppinger, C, Wohlers, H, Kayastha, K, Demmer, U, Ermler, U, Mueller, V. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.133 Å) | | Cite: | Molecular basis of the flavin-based electron-bifurcating caffeyl-CoA reductase reaction.
FEBS Lett., 592, 2018
|
|
3IQZ
 
 | | Structure of F420 dependent methylene-tetrahydromethanopterin dehydrogenase in complex with methylene-tetrahydromethanopterin | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, CALCIUM ION, F420-dependent methylenetetrahydromethanopterin dehydrogenase, ... | | Authors: | Ceh, K.E, Demmer, U, Warkentin, E, Moll, J, Thauer, R.K, Shima, S, Ermler, U. | | Deposit date: | 2009-08-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the hydride transfer mechanism in F(420)-dependent methylenetetrahydromethanopterin dehydrogenase
Biochemistry, 48, 2009
|
|
3IQE
 
 | | Structure of F420 dependent methylene-tetrahydromethanopterin dehydrogenase in complex with methylene-tetrahydromethanopterin and coenzyme F420 | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, CALCIUM ION, COENZYME F420, ... | | Authors: | Ceh, K.E, Demmer, U, Warkentin, E, Moll, J, Thauer, R.K, Shima, S, Ermler, U. | | Deposit date: | 2009-08-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the hydride transfer mechanism in F(420)-dependent methylenetetrahydromethanopterin dehydrogenase
Biochemistry, 48, 2009
|
|
3IQF
 
 | | Structure of F420 dependent methylene-tetrahydromethanopterin dehydrogenase in complex with methenyl-tetrahydromethanopterin | | Descriptor: | 1-{4-[(6S,6aR,7R)-3-amino-6,7-dimethyl-1-oxo-1,2,5,6,6a,7-hexahydro-8H-imidazo[1,5-f]pteridin-10-ium-8-yl]phenyl}-1-deoxy-5-O-{5-O-[(S)-{[(1S)-1,3-dicarboxypropyl]oxy}(hydroxy)phosphoryl]-alpha-D-ribofuranosyl}-D-ribitol, CALCIUM ION, F420-dependent methylenetetrahydromethanopterin dehydrogenase, ... | | Authors: | Ceh, K.E, Demmer, U, Warkentin, E, Moll, J, Thauer, R.K, Shima, S, Ermler, U. | | Deposit date: | 2009-08-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the hydride transfer mechanism in F(420)-dependent methylenetetrahydromethanopterin dehydrogenase
Biochemistry, 48, 2009
|
|
1Y5Y
 
 | | Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1 | | Descriptor: | CALCIUM ION, Formaldehyde-activating enzyme fae, SODIUM ION | | Authors: | Acharya, P, Goenrich, M, Hagemeier, C.H, Demmer, U, Vorholt, J.A, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-12-03 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How an enzyme binds the C1-carrier tetrahydromethanopterin: Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1
J.Biol.Chem., 280, 2005
|
|
1Y60
 
 | | Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1 with bound 5,10-methylene tetrahydromethanopterin | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Formaldehyde-activating enzyme fae | | Authors: | Acharya, P, Goenrich, M, Hagemeier, C.H, Demmer, U, Vorholt, J.A, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-12-03 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How an enzyme binds the C1-carrier tetrahydromethanopterin: Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1
J.Biol.Chem., 280, 2005
|
|
1LU9
 
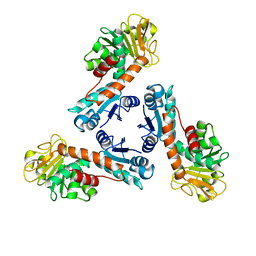 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1 | | Descriptor: | Methylene Tetrahydromethanopterin Dehydrogenase | | Authors: | Ermler, U, Hagemeier, C.H, Roth, A, Demmer, U, Grabarse, W, Warkentin, E, Vorholt, J.A. | | Deposit date: | 2002-05-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of methylene-tetrahydromethanopterin dehydrogenase from methylobacterium extorquens AM1.
Structure, 10, 2002
|
|
1LUA
 
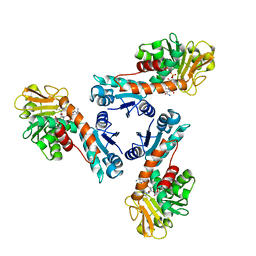 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1 complexed with NADP | | Descriptor: | Methylene Tetrahydromethanopterin Dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ermler, U, Hagemeier, C.H, Roth, A, Demmer, U, Grabarse, W, Warkentin, E, Vorholt, J.A. | | Deposit date: | 2002-05-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of methylene-tetrahydromethanopterin dehydrogenase from methylobacterium extorquens AM1.
Structure, 10, 2002
|
|
3SQG
 
 | | Crystal structure of a methyl-coenzyme M reductase purified from Black Sea mats | | Descriptor: | (17[2]S)-17[2]-methylthio-coenzyme F43, 1-THIOETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Shima, S, Krueger, M, Weinert, T, Demmer, U, Thauer, R.K, Ermler, U. | | Deposit date: | 2011-07-05 | | Release date: | 2011-11-30 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically.
Nature, 481, 2011
|
|
7PYT
 
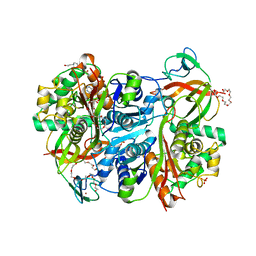 | | Benzoylsuccinyl-CoA thiolase with coenzyme A | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Benzoylsuccinyl-CoA thiolase subunit, ... | | Authors: | Ermler, U, Heider, J, Weidenweber, S, Demmer, U. | | Deposit date: | 2021-10-11 | | Release date: | 2022-04-06 | | Last modified: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Finis tolueni: a new type of thiolase with an integrated Zn-finger subunit catalyzes the final step of anaerobic toluene metabolism.
Febs J., 289, 2022
|
|
5MJI
 
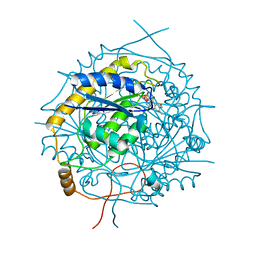 | | Crystal Structure of RosB with bound intermediate OHC-RP (8-demethyl-8-formylriboflavin-5'-phosphate) | | Descriptor: | BRAMP domain protein, [(2~{S},3~{R},4~{R})-5-[8-methanoyl-7-methyl-2,4-bis(oxidanylidene)-1~{H}-benzo[g]pteridin-10-ium-10-yl]-2,3,4-tris(oxidanyl)pentyl] dihydrogen phosphate | | Authors: | Konjik, V, Bruenle, S, Demmer, U, Vanselow, A, Sandhoff, R, Mack, M, Ermler, U. | | Deposit date: | 2016-12-01 | | Release date: | 2016-12-28 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of RosB: Insights into the Reaction Mechanism of the First Member of a Family of Flavodoxin-like Enzymes.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MLD
 
 | | Crystal Structure of RosB with bound intermediate AFP (8-demethyl-8-aminoriboflavin-5'-phosphate) | | Descriptor: | 8-demethyl-8-aminoriboflavin-5'-phosphate, BRAMP domain protein | | Authors: | Konjik, V, Bruenle, S, Demmer, U, Vanselow, A, Sandhoff, R, Mack, M, Ermler, U. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-20 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of RosB gives insights into the reaction mechanism of this first known member of a new family of flavodoxin-like enzymes
To be published
|
|
4DNX
 
 | | The structure of the ATP sulfurylase from Allochromatium vinosum in the open state | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sulfate adenylyltransferase | | Authors: | Parey, K, Demmer, U, Warkentin, E, Dahl, C, Ermler, U. | | Deposit date: | 2012-02-09 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, biochemical and genetic characterization of dissimilatory ATP sulfurylase from Allochromatium vinosum.
Plos One, 8, 2013
|
|
4F6T
 
 | | The crystal structure of the molybdenum storage protein (MoSto) from Azotobacter vinelandii loaded with various polyoxometalates | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MO(6)-O(26) Cluster, ... | | Authors: | Kowalewski, B, Poppe, J, Schneider, K, Demmer, U, Warkentin, E, Ermler, U. | | Deposit date: | 2012-05-15 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nature's Polyoxometalate Chemistry: X-ray Structure of the Mo Storage Protein Loaded with Discrete Polynuclear Mo-O Clusters.
J.Am.Chem.Soc., 134, 2012
|
|
