4AU1
 
 | | Crystal Structure of CobH (precorrin-8x methyl mutase) complexed with C5 desmethyl-HBA | | Descriptor: | DESMETHYL-HBA, PRECORRIN-8X METHYLMUTASE, SULFATE ION | | Authors: | Deery, E, Lawrence, A.D, Schroeder, S, Taylor, S.L, Seyedarabi, A, Vevodova, J, Wilson, K.S, Brown, D, Geeves, M.A, Howard, M.J, Pickersgill, R.W, Warren, M.J. | | Deposit date: | 2012-05-11 | | Release date: | 2012-09-19 | | Last modified: | 2012-11-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An Enzyme-Trap Approach Allows Isolation of Intermediates in Cobalamin Biosynthesis
Nat.Chem.Biol., 8, 2012
|
|
4FDV
 
 | | CobH from Rhodobacter capsulatus (SB1003) in complex with HBA | | Descriptor: | 3-[(1R,2R,3R,5R,6S,7R,9Z,12S,13S,14Z,17S,18S,19R)-2,13,18-tris(2-hydroxy-2-oxoethyl)-3,12,17-tris(3-hydroxy-3-oxopropyl)-3,5,8,8,13,15,18,19-octamethyl-1,2,5,6,7,12,17,22-octahydrocorrin-7-yl]propanoic acid, GLYCEROL, Precorrin-8X methylmutase | | Authors: | Pickersgill, R.W, Schroeder, S, Deery, E, Warren, M.J. | | Deposit date: | 2012-05-29 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | An enzyme-trap approach allows isolation of intermediates in cobalamin biosynthesis.
Nat.Chem.Biol., 8, 2012
|
|
4X7G
 
 | | CobK precorrin-6A reductase | | Descriptor: | 3-[(1R,2S,3S,7S,11S,16S,17R,18R,19R)-2,7,12,18-tetrakis(2-hydroxy-2-oxoethyl)-3,13-bis(3-hydroxy-3-oxopropyl)-1,2,5,7,11,17-hexamethyl-17-(3-oxidanyl-3-oxidanylidene-prop-1-enyl)-3,6,8,10,15,16,18,19,21,24-decahydrocorrin-8-yl]propanoic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Precorrin-6A reductase | | Authors: | Gu, S, Deery, E, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2014-12-09 | | Release date: | 2016-01-20 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal structure of CobK reveals strand-swapping between Rossmann-fold domains and molecular basis of the reduced precorrin product trap.
Sci Rep, 5, 2015
|
|
4MLV
 
 | | Crystal Structure of Bacillus megaterium porphobilinogen deaminase | | Descriptor: | 3-[(5S)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, ... | | Authors: | Azim, N, Deery, E, Warren, M.J, Erskine, P, Cooper, J.B, Coker, A, Wood, S.P, Akhtar, M. | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.455 Å) | | Cite: | Structural evidence for the partially oxidized dipyrromethene and dipyrromethanone forms of the cofactor of porphobilinogen deaminase: structures of the Bacillus megaterium enzyme at near-atomic resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MLQ
 
 | | Crystal structure of Bacillus megaterium porphobilinogen deaminase | | Descriptor: | 3-[(5S)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, ... | | Authors: | Azim, N, Deery, E, Warren, M.J, Erskine, P, Cooper, J.B, Coker, A, Wood, S.P, Akhtar, M. | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural evidence for the partially oxidized dipyrromethene and dipyrromethanone forms of the cofactor of porphobilinogen deaminase: structures of the Bacillus megaterium enzyme at near-atomic resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CCS
 
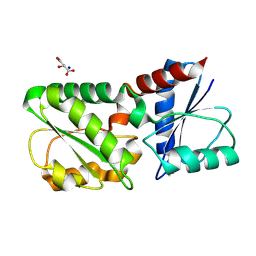 | | The structure of CbiX, the terminal Enzyme for Biosynthesis of Siroheme in Denitrifying Bacteria | | Descriptor: | 1,2-ETHANEDIOL, CBIX, D-MALATE, ... | | Authors: | Bali, S, Rollauer, S.E, Roversi, P, Raux-Deery, E, Lea, S.M, Warren, M.J, Ferguson, S.J. | | Deposit date: | 2013-10-25 | | Release date: | 2014-04-09 | | Last modified: | 2015-09-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Characterization of the 'Missing' Terminal Enzyme for Siroheme Biosynthesis in Alpha-Proteobacteria.
Mol.Microbiol., 92, 2014
|
|
2W6K
 
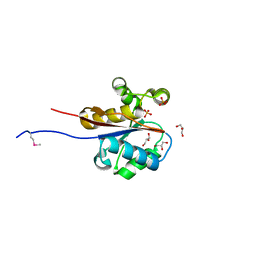 | | The crystal structure at 1.7 A resolution of CobE, a protein from the cobalamin (vitamin B12) biosynthetic pathway | | Descriptor: | COBE, GLYCEROL, SULFATE ION | | Authors: | Vevodova, J, Smith, D, McGoldrick, H, Deery, E, Murzin, A.G, Warren, M.J, Wilson, K.S. | | Deposit date: | 2008-12-18 | | Release date: | 2008-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure at 1.7 A Resolution of Cobe, a Protein from the Cobalamin (Vitamin B12) Biosynthetic Pathway
To be Published
|
|
2W6L
 
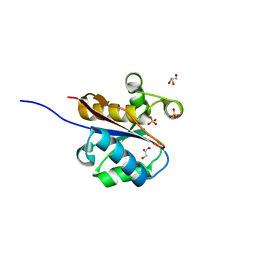 | | The crystal structure at 1.7 A resolution of CobE, a protein from the cobalamin (vitamin B12) biosynthetic pathway | | Descriptor: | COBE, GLYCEROL, SULFATE ION | | Authors: | Vevodova, J, Smith, D, McGoldrick, H, Deery, E, Murzin, A.G, Warren, M.J, Wilson, K.S. | | Deposit date: | 2008-12-18 | | Release date: | 2008-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Crystal Structure at 1.7 A Resolution of Cobe, a Protein from the Cobalamin (Vitamin B12) Biosynthetic Pathway
To be Published
|
|
2XWP
 
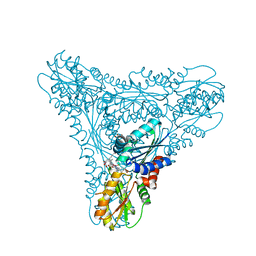 | | ANAEROBIC COBALT CHELATASE (CbiK) FROM SALMONELLA TYPHIMURIUM IN COMPLEX WITH METALATED TETRAPYRROLE | | Descriptor: | COBALT SIROHYDROCHLORIN, GLYCEROL, SIROHYDROCHLORIN COBALTOCHELATASE | | Authors: | Ladakis, D, Brindley, A.A, Deery, E, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2010-11-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution in a Family of Chelatases Facilitated by the Introduction of Active Site Asymmetry and Protein Oligomerization.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XWS
 
 | | ANAEROBIC COBALT CHELATASE (CbiX) FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | SIROHYDROCHLORIN COBALTOCHELATASE | | Authors: | Romao, C.V, Ladakis, D, Lobo, S.A.L, Carrondo, M.A, Brindley, A.A, Deery, E, Matias, P.M, Pickersgill, R.W, Saraiva, L.M, Warren, M.J. | | Deposit date: | 2010-11-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution in a Family of Chelatases Facilitated by the Introduction of Active Site Asymmetry and Protein Oligomerization.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XWQ
 
 | | Anaerobic cobalt chelatase from Archeaoglobus fulgidus (CbiX) in complex with metalated sirohydrochlorin product | | Descriptor: | COBALT SIROHYDROCHLORIN, SIROHYDROCHLORIN COBALTOCHELATASE | | Authors: | Ladakis, D, Brindley, A.A, Deery, E, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2010-11-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Evolution in a Family of Chelatases Facilitated by the Introduction of Active Site Asymmetry and Protein Oligomerization.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NJR
 
 | |
4UN1
 
 | | Sirohaem decarboxylase AhbA/B - an enzyme with structural homology to the Lrp/AsnC transcription factor family that is part of the alternative haem biosynthesis pathway. | | Descriptor: | 12,18-DIDECARBOXY-SIROHEME, PUTATIVE TRANSCRIPTIONAL REGULATOR, ASNC FAMILY | | Authors: | Palmer, D.J, Brown, D.G, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2014-05-23 | | Release date: | 2014-06-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Structure, Function and Properties of Sirohaem Decarboxylase - an Enzyme with Structural Homology to a Transcription Factor Family that is Part of the Alternative Haem Biosynthesis Pathway.
Mol.Microbiol., 93, 2014
|
|
3CB0
 
 | | CobR | | Descriptor: | 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE, FLAVIN MONONUCLEOTIDE | | Authors: | Lawrence, A.D, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2008-02-21 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification, characterization, and structure/function analysis of a corrin reductase involved in adenosylcobalamin biosynthesis
J.Biol.Chem., 283, 2008
|
|
5C4R
 
 | | CobK precorrin-6A reductase | | Descriptor: | Precorrin-6A reductase | | Authors: | Gu, S, Pickersgill, R.W. | | Deposit date: | 2015-06-18 | | Release date: | 2016-11-16 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Crystal structure of CobK reveals strand-swapping between Rossmann-fold domains and molecular basis of the reduced precorrin product trap.
Sci Rep, 5, 2015
|
|
2DJ5
 
 | | Crystal Structure of the vitamin B12 biosynthetic cobaltochelatase, CbiXS, from Archaeoglobus fulgidus | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sirohydrochlorin cobaltochelatase | | Authors: | Yin, J, Cherney, M.M, James, M.N.G. | | Deposit date: | 2006-03-31 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the Vitamin B(12) Biosynthetic Cobaltochelatase, CbiX (S), from Archaeoglobus Fulgidus
J.STRUCT.FUNCT.GENOM., 7, 2006
|
|
2QBU
 
 | | Crystal structure of Methanothermobacter thermautotrophicus CbiL | | Descriptor: | Precorrin-2 methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Frank, S, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidation of substrate specificity in the cobalamin (vitamin B12) biosynthetic methyltransferases. Structure and function of the C20 methyltransferase (CbiL) from Methanothermobacter thermautotrophicus.
J.Biol.Chem., 282, 2007
|
|
4CZD
 
 | |
2XVX
 
 | | Cobalt chelatase CbiK (periplasmatic) from Desulvobrio vulgaris Hildenborough (Native) | | Descriptor: | CARBON DIOXIDE, CHELATASE, PUTATIVE, ... | | Authors: | Romao, C.V, Lobo, S.A.L, Carrondo, M.A, Saraiva, L.M, Matias, P.M. | | Deposit date: | 2010-10-28 | | Release date: | 2010-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution in a Family of Chelatases Facilitated by the Introduction of Active Site Asymmetry and Protein Oligomerization.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XVZ
 
 | | Cobalt chelatase CbiK (periplasmatic) from Desulvobrio vulgaris Hildenborough (co-crystallized with cobalt) | | Descriptor: | CHELATASE, PUTATIVE, CHLORIDE ION, ... | | Authors: | Romao, C.V, Lobo, S.A.L, Carrondo, M.A, Saraiva, L.M, Matias, P.M. | | Deposit date: | 2010-10-28 | | Release date: | 2010-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution in a Family of Chelatases Facilitated by the Introduction of Active Site Asymmetry and Protein Oligomerization.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
