2L8T
 
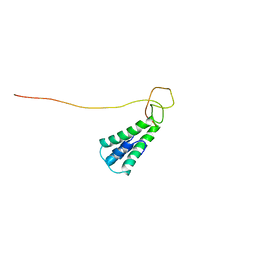 | | Staphylococcus aureus pathogenicity island 1 protein gp6, an internal scaffold in size determination | | Descriptor: | Transposon Tn557 toxic shock syndrome toxin-1 | | Authors: | Dearborn, A.D, Spilman, M.S, Damle, P.K, Chang, J.R, Monroe, E.B, Saad, J.S, Christie, G.E, Dokland, T. | | Deposit date: | 2011-01-24 | | Release date: | 2011-08-17 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | The Staphylococcus aureus Pathogenicity Island 1 Protein gp6 Functions as an Internal Scaffold during Capsid Size Determination.
J.Mol.Biol., 412, 2011
|
|
6B0X
 
 | |
6B23
 
 | |
6CF2
 
 | | Crystal structure of HIV-1 Rev (residues 1-93)-RNA aptamer complex | | Descriptor: | Anti-Rev Antibody, heavy chain, light chain, ... | | Authors: | Eren, E, Dearborn, A.D, Wingfield, P.T. | | Deposit date: | 2018-02-13 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of an RNA Aptamer that Can Inhibit HIV-1 by Blocking Rev-Cognate RNA (RRE) Binding and Rev-Rev Association.
Structure, 26, 2018
|
|
2P4V
 
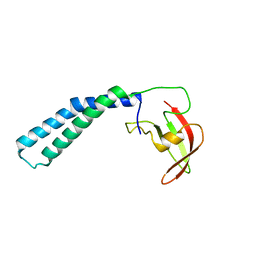 | | Crystal structure of the transcript cleavage factor, GreB at 2.6A resolution | | Descriptor: | Transcription elongation factor greB | | Authors: | Vassylyeva, M.N, Svetlov, V, Dearborn, A.D, Klyuyev, S, Artsimovitch, I, Vassylyev, D.G. | | Deposit date: | 2007-03-13 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The carboxy-terminal coiled-coil of the RNA polymerase beta'-subunit is the main binding site for Gre factors.
Embo Rep., 8, 2007
|
|
8DK6
 
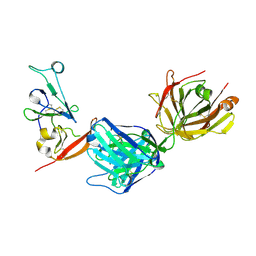 | | Structure of hepatitis C virus envelope N-terminal truncated glycoprotein 2 (E2) (residues 456-713) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, 2A12 Fab light chain, ... | | Authors: | Kumar, A, Rohe, T, Elrod, E.J, Khan, A.G, Dearborn, A.D, Kissinger, R, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Regions of hepatitis C virus E2 required for membrane association.
Nat Commun, 14, 2023
|
|
7MWW
 
 | | Structure of hepatitis C virus envelope full-length glycoprotein 2 (eE2) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, ... | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
7MWS
 
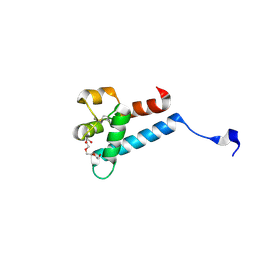 | | Crystal structure of tamarin CD81 large extracellular loop | | Descriptor: | CD81 protein, GLYCEROL, TETRAETHYLENE GLYCOL | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
7MWX
 
 | | Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2 with tamarin CD81 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy Chain, ... | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
6C22
 
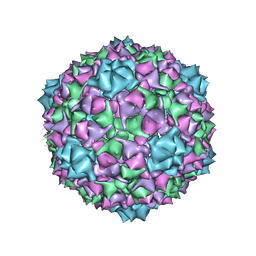 | |
6C21
 
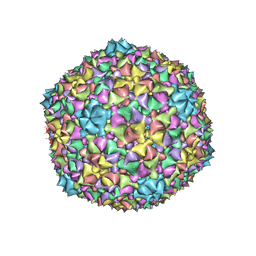 | |
6V8I
 
 | |
6CWT
 
 | |
6CVK
 
 | |
6CWD
 
 | |
