5B81
 
 | |
3FW0
 
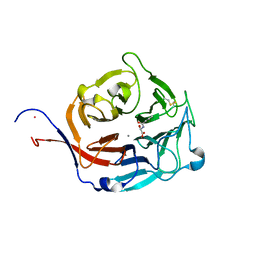 | | Structure of Peptidyl-alpha-hydroxyglycine alpha-Amidating Lyase (PAL) bound to alpha-hydroxyhippuric acid (non-peptidic substrate) | | Descriptor: | (2S)-hydroxy[(phenylcarbonyl)amino]ethanoic acid, CALCIUM ION, MERCURY (II) ION, ... | | Authors: | Chufan, E.E, De, M, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2009-01-16 | | Release date: | 2009-09-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme.
Structure, 17, 2009
|
|
3FVZ
 
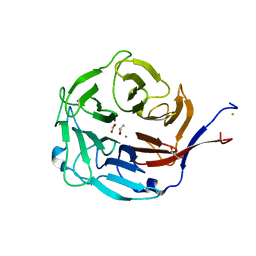 | | Structure of Peptidyl-alpha-hydroxyglycine alpha-Amidating Lyase (PAL) | | Descriptor: | ACETATE ION, CALCIUM ION, FE (III) ION, ... | | Authors: | Chufan, E.E, De, M, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2009-01-16 | | Release date: | 2009-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme.
Structure, 17, 2009
|
|
