3X2W
 
 | | Michaelis complex of cAMP-dependent Protein Kinase Catalytic Subunit | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | Authors: | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
3X2V
 
 | | Michaelis-like complex of cAMP-dependent Protein Kinase Catalytic Subunit | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | Authors: | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
3X2U
 
 | | Michaelis-like initial complex of cAMP-dependent Protein Kinase Catalytic Subunit. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Substrate Peptide, ... | | Authors: | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
5YRS
 
 | | X-ray Snapshot of HIV-1 Protease in Action: Observation of Tetrahedral Intermediate and Its SIHB with Catalytic Aspartate | | Descriptor: | PROTEASE, RT-RH oligopeprtide | | Authors: | Das, A, Mahale, S, Prashar, V, Bihani, S, Ferrer, J.-L, Hosur, M.V. | | Deposit date: | 2017-11-10 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray snapshot of HIV-1 protease in action: observation of tetrahedral intermediate and short ionic hydrogen bond SIHB with catalytic aspartate.
J. Am. Chem. Soc., 132, 2010
|
|
5DN7
 
 | | Crescerin uses a TOG domain array to regulate microtubules in the primary cilium | | Descriptor: | Protein FAM179B | | Authors: | Das, A, Dickinson, D.J, Wood, C.C, Goldstein, B, Slep, K.C. | | Deposit date: | 2015-09-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crescerin uses a TOG domain array to regulate microtubules in the primary cilium.
Mol.Biol.Cell, 26, 2015
|
|
7L3L
 
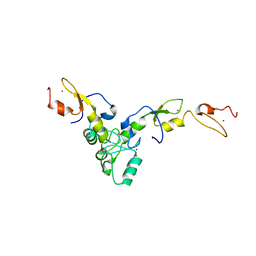 | | Structure of TRAF5 and TRAF6 RING Hetero dimer | | Descriptor: | TNF receptor-associated factor 5, TNF receptor-associated factor 6, ZINC ION | | Authors: | Das, A, Middleton, A.J, Padala, P, Day, C.L. | | Deposit date: | 2020-12-17 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure and ubiquitin binding properties of TRAF RING heterodimers.
J.Mol.Biol., 2021
|
|
6WC0
 
 | |
6WBR
 
 | |
2NPH
 
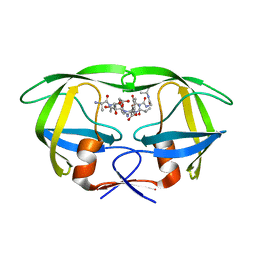 | | Crystal structure of HIV1 protease in situ product complex | | Descriptor: | PROTEASE RETROPEPSIN, pentapeptide fragment, tetrapeptide fragment | | Authors: | Hosur, M.V, Das, A, Prashar, V. | | Deposit date: | 2006-10-27 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of HIV-1 protease in situ product complex and observation of a low-barrier hydrogen bond between catalytic aspartates
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6KMP
 
 | | 100K X-ray structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate mimic KVS-1 | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, Protease | | Authors: | Das, A, Kovalevsky, A. | | Deposit date: | 2019-07-31 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
4O22
 
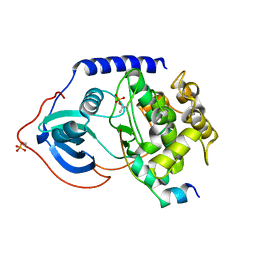 | | Binary complex of metal-free PKAc with SP20. | | Descriptor: | Phosphorylated peptide pSP20., cAMP-dependent protein kinase catalytic subunit alpha. | | Authors: | Das, A, Kovalevsky, A.Y, Gerlits, O, Langan, P, Heller, W.T, Keshwani, M, Taylor, S.S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-Free cAMP-Dependent Protein Kinase Can Catalyze Phosphoryl Transfer.
Biochemistry, 53, 2014
|
|
4O21
 
 | | Product complex of metal-free PKAc, ATP-gamma-S and SP20. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thio-phosphorylated peptide pSP20, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Das, A, Kovalevsky, A.Y, Gerlits, O, Langan, P, Heller, W.T, Keshwani, M, Taylor, S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Metal-Free cAMP-Dependent Protein Kinase Can Catalyze Phosphoryl Transfer.
Biochemistry, 53, 2014
|
|
8D2L
 
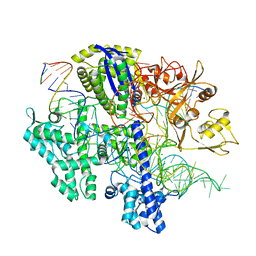 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Cleavage Intermediate 1) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*GP*A)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9
Nat Catal, 6, 2023
|
|
8D2K
 
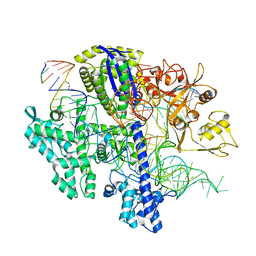 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Cleavage Intermediate 2) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*GP*A)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9
Nat Catal, 6, 2023
|
|
8D2N
 
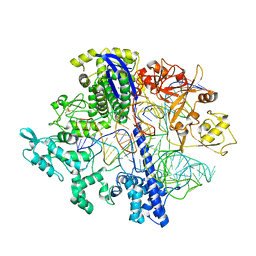 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Pre-cleavage) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*TP*AP*CP*AP*CP*CP*AP*AP*GP*CP*T)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9
Nat Catal, 6, 2023
|
|
8D2Q
 
 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 1) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*TP*AP*CP*AP*CP*CP*AP*AP*GP*CP*T)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9
Nat Catal, 6, 2023
|
|
8D2P
 
 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Target bound) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA target strand (5'-D(P*CP*CP*AP*GP*GP*AP*TP*CP*TP*TP*GP*CP*CP*AP*TP*CP*CP*TP*AP*CP*CP*TP*CP*T)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9
Nat Catal, 6, 2023
|
|
8D2O
 
 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 2) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*TP*AP*CP*AP*CP*CP*AP*AP*GP*CP*T)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9
Nat Catal, 6, 2023
|
|
8FAI
 
 | | Cryo-EM structure of the Agrobacterium T-pilus | | Descriptor: | (7Z,19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacos-7-en-19-yl (9Z)-octadec-9-enoate, Protein virB2 | | Authors: | Kreida, S, Narita, A, Johnson, M.D, Tocheva, E.I, Das, A, Jensen, G.J, Ghosal, D. | | Deposit date: | 2022-11-27 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the Agrobacterium tumefaciens T4SS-associated T-pilus reveals stoichiometric protein-phospholipid assembly.
Structure, 31, 2023
|
|
6PU8
 
 | | Room temperature X-ray structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate of keto-darunavir | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3,3-dihydroxy-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Kovalevsky, A, Das, A. | | Deposit date: | 2019-07-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
6PTP
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate mimic KVS-1 | | Descriptor: | HIV-1 Protease, N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide | | Authors: | Kovalevsky, A, Das, A. | | Deposit date: | 2019-07-16 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
4XW6
 
 | | X-ray structure of PKAc with ADP, free phosphate ion, CP20, magnesium ions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | Deposit date: | 2015-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
4XW5
 
 | | X-ray structure of PKAc with ATP, CP20, calcium ions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | Deposit date: | 2015-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
4XW4
 
 | | X-ray structure of PKAc with AMPPNP, SP20, calcium ions | | Descriptor: | CALCIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | Deposit date: | 2015-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2015-07-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
1JO1
 
 | | N7-Guanine Adduct of 2,7-diaminomitosene with DNA | | Descriptor: | 5'-D(*GP*TP*GP*(DAJ)GP*TP*AP*TP*AP*CP*CP*AP*C)-3', DECARBAMOYL-2,7-DIAMINOMITOSENE | | Authors: | Subramaniam, G, Paz, M.M, Kumar, G.S, Das, A, Palom, Y, Clement, C.C, Patel, D.J, Tomasz, M. | | Deposit date: | 2001-07-26 | | Release date: | 2001-09-12 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a guanine-N7-linked complex of the mitomycin C metabolite 2,7-diaminomitosene and DNA. Basis of sequence selectivity.
Biochemistry, 40, 2001
|
|
