2GJH
 
 | |
2A3J
 
 | | Structure of URNdesign, a complete computational redesign of human U1A protein | | Descriptor: | U1 small nuclear ribonucleoprotein A | | Authors: | Varani, G, Dobson, N, Dantas, G, Baker, D. | | Deposit date: | 2005-06-24 | | Release date: | 2006-06-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Structural Validation of the Computational Redesign of Human U1A Protein
Structure, 14, 2006
|
|
1QYS
 
 | | Crystal structure of Top7: A computationally designed protein with a novel fold | | Descriptor: | TOP7 | | Authors: | Kuhlman, B, Dantas, G, Ireton, G.C, Varani, G, Stoddard, B.L, Baker, D. | | Deposit date: | 2003-09-11 | | Release date: | 2003-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of a Novel Globular Protein Fold with Atomic-Level Accuracy
Science, 302, 2003
|
|
2GJF
 
 | |
1VJQ
 
 | |
5HT0
 
 | | Crystal structure of an Antibiotic_NAT family aminoglycoside acetyltransferase HMB0038 from an uncultured soil metagenomic sample in complex with coenzyme A | | Descriptor: | Aminoglycoside acetyltransferase HMB0005, COENZYME A, SULFATE ION | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
8ER1
 
 | | X-ray crystal structure of Tet(X6) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2022-10-11 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of anhydrotetracycline-bound Tet(X6) reveals the mechanism for inhibition of type 1 tetracycline destructases.
Commun Biol, 6, 2023
|
|
8ER0
 
 | | X-ray crystal structure of Tet(X6) bound to anhydrotetracycline | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2022-10-11 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of anhydrotetracycline-bound Tet(X6) reveals the mechanism for inhibition of type 1 tetracycline destructases.
Commun Biol, 6, 2023
|
|
6WG9
 
 | | Crystal structure of tetracycline destructase Tet(X7) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Tetracycline destructase Tet(X7) | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2020-04-05 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Tetracycline-inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance.
Commun Biol, 3, 2020
|
|
5F49
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with malonyl-coenzyme A | | Descriptor: | COENZYME A, MAGNESIUM ION, MALONYL-COENZYME A, ... | | Authors: | Xu, Z, Skarina, T, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
5F46
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample, apoenzyme form | | Descriptor: | CHLORIDE ION, aminoglycoside acetyltransferase meta-AAC0020 | | Authors: | Xu, Z, Skarina, T, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
5F48
 
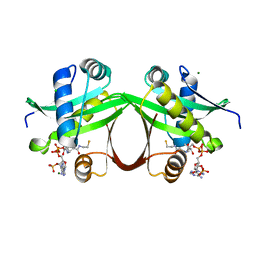 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with coenzyme A | | Descriptor: | CHLORIDE ION, COENZYME A, MAGNESIUM ION, ... | | Authors: | Xu, Z, Skarina, T, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
5F47
 
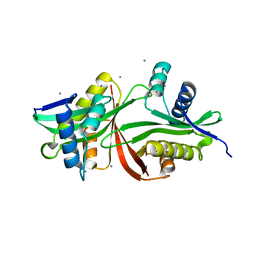 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with trehalose | | Descriptor: | CALCIUM ION, CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Xu, Z, Skarina, T, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
7KES
 
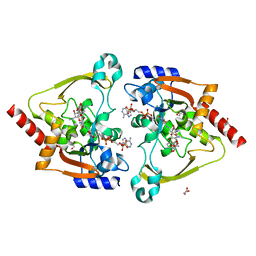 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in complex with apramycin and CoA | | Descriptor: | APRAMYCIN, Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-12 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
7LAP
 
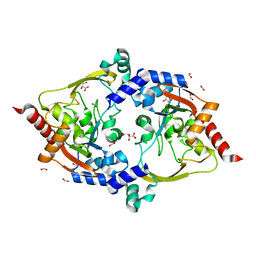 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Xa | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, D(-)-TARTARIC ACID, ... | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-06 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
7LAO
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IIb | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aminoglycoside N(3)-acetyltransferase III, MAGNESIUM ION | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-06 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6MN5
 
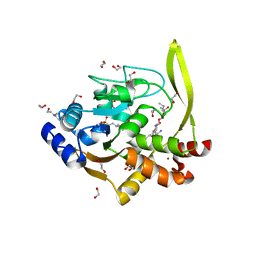 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IVa, H154A mutant, in complex with gentamicin C1A | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6MMZ
 
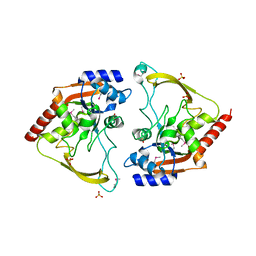 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H29A mutant apoenzyme | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, SULFATE ION | | Authors: | Stogios, P.J, Skarina, T, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6MN4
 
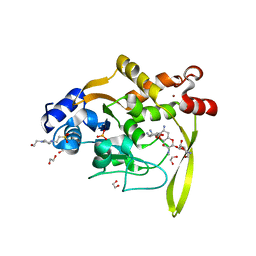 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IVa, H154A mutant, in complex with apramycin | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, APRAMYCIN, ... | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6MN3
 
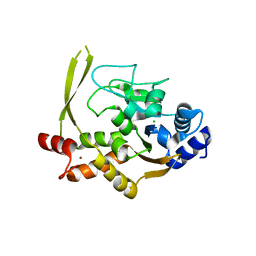 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IVa, apoenzyme | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, AAC(3)-IVa, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6MN0
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H168A mutant in complex with acetyl-CoA | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETYL COENZYME *A, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Zu, X, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
5TUI
 
 | | Crystal structure of tetracycline destructase Tet(50) in complex with chlortetracycline | | Descriptor: | 7-CHLOROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUE
 
 | | Crystal structure of tetracycline destructase Tet(50) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Tetracycline destructase Tet(50) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUK
 
 | | Crystal structure of tetracycline destructase Tet(51) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Tetracycline destructase Tet(51) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUM
 
 | | Crystal structure of tetracycline destructase Tet(56) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, tetracycline destructase Tet(56) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
