3TE0
 
 | | Crystal structure of HSC K148E | | Descriptor: | formate/nitrite transporter, octyl beta-D-glucopyranoside | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3TDO
 
 | | Crystal structure of HSC at pH 9.0 | | Descriptor: | Putative formate/nitrite transporter, TETRAETHYLENE GLYCOL, octyl beta-D-glucopyranoside | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3TDX
 
 | | Crystal structure of HSC L82V | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, formate/nitrite transporter, ... | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3TE2
 
 | | Crystal structure of HSC K16S | | Descriptor: | TETRAETHYLENE GLYCOL, formate/nitrite transporter, octyl beta-D-glucopyranoside | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3TDR
 
 | | Crystal structure of HSC at pH 7.5 | | Descriptor: | TETRAETHYLENE GLYCOL, formate/nitrite transporter | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3TDS
 
 | | Crystal structure of HSC F194I | | Descriptor: | TETRAETHYLENE GLYCOL, formate/nitrite transporter, octyl beta-D-glucopyranoside | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3TE1
 
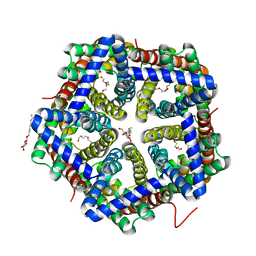 | | Crystal structure of HSC T84A | | Descriptor: | TETRAETHYLENE GLYCOL, formate/nitrite transporter, octyl beta-D-glucopyranoside | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3TDP
 
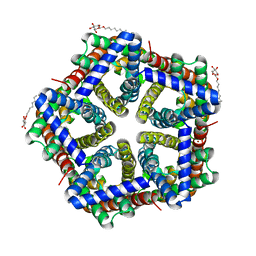 | | Crystal structure of HSC at pH 4.5 | | Descriptor: | ZINC ION, formate/nitrite transporter, octyl beta-D-glucopyranoside | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
1Z3I
 
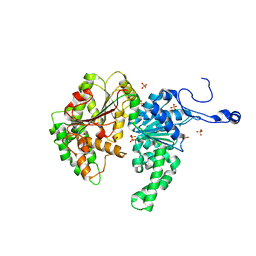 | | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | | Descriptor: | SULFATE ION, ZINC ION, similar to RAD54-like | | Authors: | Thoma, N.H, Czyzewski, B.K, Alexeev, A.A, Mazin, A.V, Kowalczykowski, S.C, Pavletich, N.P. | | Deposit date: | 2005-03-12 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54.
Nat.Struct.Mol.Biol., 12, 2005
|
|
3EI2
 
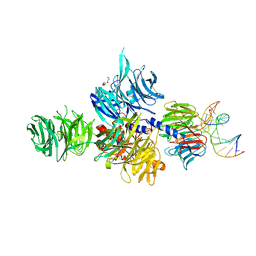 | | Structure of hsDDB1-drDDB2 bound to a 16 bp abasic site containing DNA-duplex | | Descriptor: | 5'-D(*DAP*DAP*DAP*DTP*DGP*DAP*DAP*DTP*(3DR)P*DAP*DAP*DGP*DCP*DAP*DGP*DG)-3', 5'-D(*DCP*DCP*DTP*DGP*DCP*DTP*DTP*DTP*DAP*DTP*DTP*DCP*DAP*DTP*DTP*DT)-3', DNA damage-binding protein 1, ... | | Authors: | Scrima, A, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
3EI1
 
 | |
3EI4
 
 | | Structure of the hsDDB1-hsDDB2 complex | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2 | | Authors: | Scrima, A, Pavletich, N.P, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
3EI3
 
 | | Structure of the hsDDB1-drDDB2 complex | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, TETRAETHYLENE GLYCOL | | Authors: | Scrima, A, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
