1SU9
 
 | |
1ST9
 
 | | Crystal Structure of a Soluble Domain of ResA in the Oxidised Form | | Descriptor: | 1,2-ETHANEDIOL, Thiol-disulfide oxidoreductase resA | | Authors: | Crow, A, Acheson, R.M, Le Brun, N.E, Oubrie, A. | | Deposit date: | 2004-03-25 | | Release date: | 2004-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of Redox-coupled Protein Substrate Selection by the Cytochrome c Biosynthesis Protein ResA.
J.Biol.Chem., 279, 2004
|
|
5LIL
 
 | | Structure of Aggregatibacter actinomycetemcomitans MacB bound to ATPyS (P21) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Macrolide export ATP-binding/permease protein MacB | | Authors: | Crow, A. | | Deposit date: | 2016-07-15 | | Release date: | 2017-11-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LJ8
 
 | | Structure of the E. coli MacB periplasmic domain (P21) | | Descriptor: | Macrolide export ATP-binding/permease protein MacB | | Authors: | Crow, A. | | Deposit date: | 2016-07-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LJ6
 
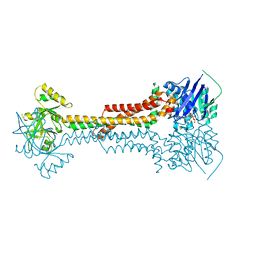 | |
5LJ9
 
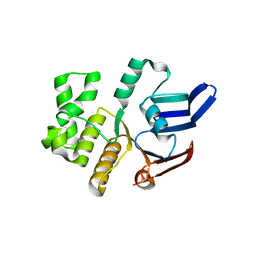 | | Structure of the E. coli MacB ABC domain (C2221) | | Descriptor: | Macrolide export ATP-binding/permease protein MacB | | Authors: | Crow, A. | | Deposit date: | 2016-07-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LJ7
 
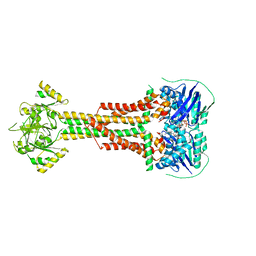 | | Structure of Aggregatibacter actinomycetemcomitans MacB bound to ATP (P21) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Macrolide export ATP-binding/permease protein MacB | | Authors: | Crow, A. | | Deposit date: | 2016-07-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LJA
 
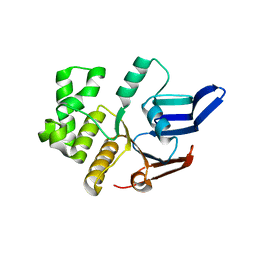 | | Structure of the E. coli MacB ABC domain (P6122) | | Descriptor: | Macrolide export ATP-binding/permease protein MacB | | Authors: | Crow, A. | | Deposit date: | 2016-07-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4XYD
 
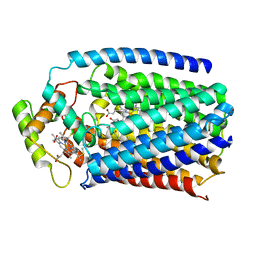 | | Nitric oxide reductase from Roseobacter denitrificans (RdNOR) | | Descriptor: | CALCIUM ION, COPPER (II) ION, FE (III) ION, ... | | Authors: | Crow, A. | | Deposit date: | 2015-02-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the Membrane-intrinsic Nitric Oxide Reductase from Roseobacter denitrificans.
Biochemistry, 55, 2016
|
|
4F8C
 
 | |
4FBJ
 
 | |
3GQJ
 
 | |
3E1P
 
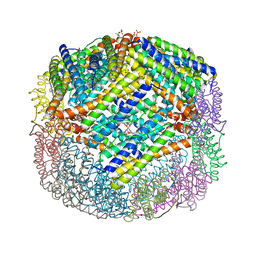 | | Crystal structure of E. coli Bacterioferritin (BFR) in which the Ferroxidase centre is inhibited with ZN(II) and high occupancy iron is bound within the cavity. | | Descriptor: | BACTERIOFERRITIN, FE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Crow, A, Lawson, T, Lewin, A, Moore, G.R, Le Brun, N. | | Deposit date: | 2008-08-04 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for iron mineralization by bacterioferritin
J.Am.Chem.Soc., 131, 2009
|
|
3E1L
 
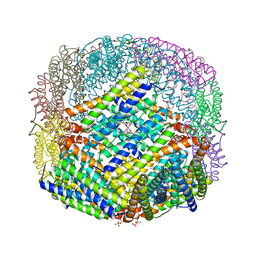 | | Crystal structure of E. coli Bacterioferritin (BFR) soaked in phosphate with an alternative conformation of the unoccupied Ferroxidase centre (APO-BFR II). | | Descriptor: | BACTERIOFERRITIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Crow, A, Lawson, T, Lewin, A, Moore, G.R, Le Brun, N. | | Deposit date: | 2008-08-04 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for iron mineralization by bacterioferritin
J.Am.Chem.Soc., 131, 2009
|
|
3E1N
 
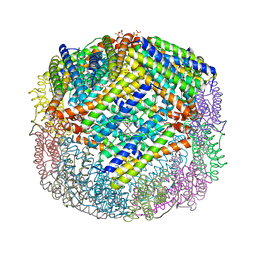 | | Crystal structure of E. coli Bacterioferritin (BFR) after a 65 minute (aerobic) exposure to FE(II) revealing a possible MU-OXO bridge/MU-Hydroxy bridged DIIRON intermediate at the ferroxidase centre. (FE(III)-O-FE(III)-BFR). | | Descriptor: | BACTERIOFERRITIN, FE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Crow, A, Lawson, T, Lewin, A, Moore, G.R, Le Brun, N. | | Deposit date: | 2008-08-04 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for iron mineralization by bacterioferritin
J.Am.Chem.Soc., 131, 2009
|
|
3E1O
 
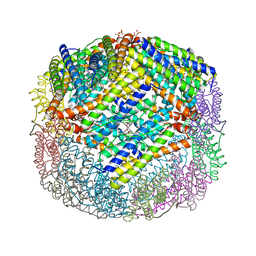 | | Crystal structure of E. coli Bacterioferritin (BFR) with two ZN(II) ION sites at the Ferroxidase centre (ZN-BFR). | | Descriptor: | BACTERIOFERRITIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Crow, A, Lawson, T, Lewin, A, Moore, G.R, Le Brun, N. | | Deposit date: | 2008-08-04 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for iron mineralization by bacterioferritin
J.Am.Chem.Soc., 131, 2009
|
|
3E1J
 
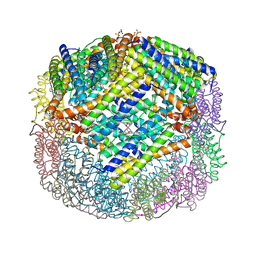 | | Crystal structure of E. coli Bacterioferritin (BFR) with an unoccupied ferroxidase centre (APO-BFR). | | Descriptor: | Bacterioferritin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Crow, A, Lawson, T, Lewin, A, Moore, G.R, Le Brun, N. | | Deposit date: | 2008-08-04 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for iron mineralization by bacterioferritin
J.Am.Chem.Soc., 131, 2009
|
|
3E1M
 
 | | Crystal structure of E. coli Bacterioferritin (BFR) obtained after soaking APO-BFR crystals for 2.5 minutes in FE2+ (2.5M FE(II)-BFR) | | Descriptor: | BACTERIOFERRITIN, FE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Crow, A, Lawson, T, Lewin, A, Moore, G.R, Le Brun, N. | | Deposit date: | 2008-08-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for iron mineralization by bacterioferritin
J.Am.Chem.Soc., 131, 2009
|
|
3GHA
 
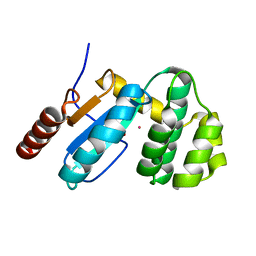 | | Crystal Structure of ETDA-treated BdbD (Reduced) | | Descriptor: | 1,2-ETHANEDIOL, Disulfide bond formation protein D, UNKNOWN ATOM OR ION | | Authors: | Crow, A, Lewin, A, Hederstedt, L, Le-Brun, N.E. | | Deposit date: | 2009-03-03 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure and Biophysical Properties of Bacillus subtilis BdbD: AN OXIDIZING THIOL:DISULFIDE OXIDOREDUCTASE CONTAINING A NOVEL METAL SITE
J.Biol.Chem., 284, 2009
|
|
3GH9
 
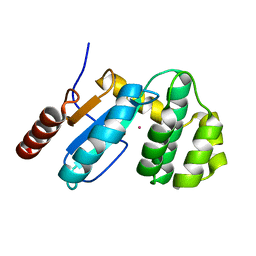 | | Crystal structure of EDTA-treated BdbD (Oxidised) | | Descriptor: | 1,2-ETHANEDIOL, Disulfide bond formation protein D, UNKNOWN ATOM OR ION | | Authors: | Crow, A, Lewin, A, Hederstedt, L, Le-Brun, N.E. | | Deposit date: | 2009-03-03 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structure and Biophysical Properties of Bacillus subtilis BdbD: AN OXIDIZING THIOL:DISULFIDE OXIDOREDUCTASE CONTAINING A NOVEL METAL SITE
J.Biol.Chem., 284, 2009
|
|
3EU4
 
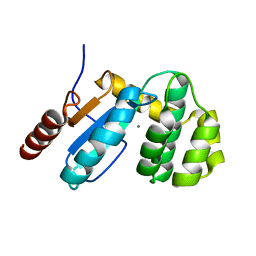 | | Crystal Structure of BdbD from Bacillus subtilis (oxidised) | | Descriptor: | BdbD, CALCIUM ION | | Authors: | Crow, A, Moller, M.C, Hederstedt, L, Le Brun, N. | | Deposit date: | 2008-10-09 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure and Biophysical Properties of Bacillus subtilis BdbD: AN OXIDIZING THIOL:DISULFIDE OXIDOREDUCTASE CONTAINING A NOVEL METAL SITE
J.Biol.Chem., 284, 2009
|
|
3EU3
 
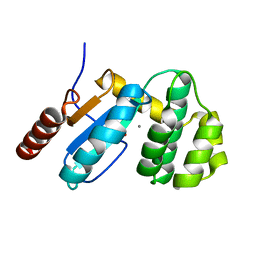 | | Crystal Structure of BdbD from Bacillus subtilis (reduced) | | Descriptor: | 1,2-ETHANEDIOL, BdbD, CALCIUM ION | | Authors: | Crow, A, Moller, M.C, Hederstedt, L, Le Brun, N.E. | | Deposit date: | 2008-10-09 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure and Biophysical Properties of Bacillus subtilis BdbD: AN OXIDIZING THIOL:DISULFIDE OXIDOREDUCTASE CONTAINING A NOVEL METAL SITE
J.Biol.Chem., 284, 2009
|
|
3ERW
 
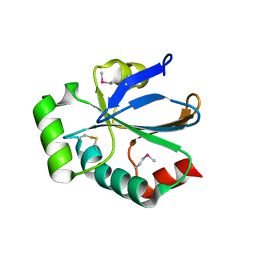 | | Crystal Structure of StoA from Bacillus subtilis | | Descriptor: | Sporulation thiol-disulfide oxidoreductase A | | Authors: | Crow, A, Liu, Y, Moller, M.C, Le Brun, N.E, Hederstedt, L. | | Deposit date: | 2008-10-03 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Functional Properties of Bacillus subtilis Endospore Biogenesis Factor StoA
J.Biol.Chem., 284, 2009
|
|
3GQM
 
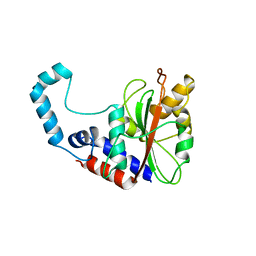 | |
4WHN
 
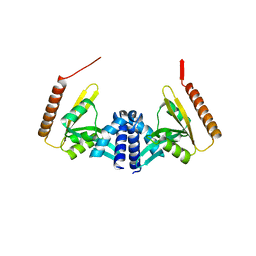 | | Structure of toxin-activating acyltransferase (TAAT) | | Descriptor: | ApxC, CITRIC ACID | | Authors: | Crow, A, Greene, N.P, Hughes, C, Koronakis, V. | | Deposit date: | 2014-09-23 | | Release date: | 2015-06-03 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of a bacterial toxin-activating acyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
