1YRS
 
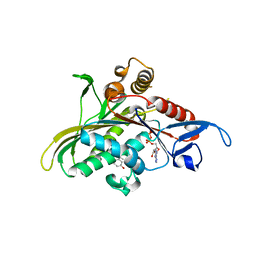 | | Crystal structure of KSP in complex with inhibitor 1 | | Descriptor: | 3-[(5S)-1-ACETYL-3-(2-CHLOROPHENYL)-4,5-DIHYDRO-1H-PYRAZOL-5-YL]PHENOL, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Cox, C.D, Breslin, M.J, Mariano, B.J, Coleman, P.J, Buser, C.A, Walsh, E.S, Hamilton, K, Kohl, N.E, Torrent, M, Yan, Y, Kuo, L.C, Hartman, G.D. | | Deposit date: | 2005-02-04 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 1: The discovery of 3,5-diaryl-4,5-dihydropyrazoles as potent and selective inhibitors of the mitotic kinesin KSP
BIOORG.MED.CHEM.LETT., 15, 2005
|
|
8IMZ
 
 | | Cryo-EM structure of mouse Piezo1-MDFIC complex (composite map) | | Descriptor: | MyoD family inhibitor domain-containing protein, Piezo-type mechanosensitive ion channel component 1 | | Authors: | Zhou, Z, Ma, X, Lin, Y, Cheng, D, Bavi, N, Li, J.V, Sutton, D, Yao, M, Harvey, N, Corry, B, Zhang, Y, Cox, C.D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | MyoD-family inhibitor proteins act as auxiliary subunits of Piezo channels.
Science, 381, 2023
|
|
6VYM
 
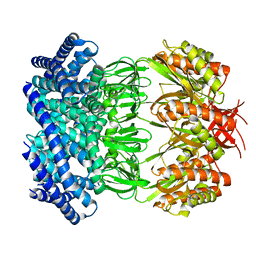 | | Cryo-EM structure of mechanosensitive channel MscS in PC-18:1 nanodiscs treated with beta-cyclodextran | | Descriptor: | Mechanosensitive channel MscS | | Authors: | Zhang, Y, Daday, C, Gu, R, Cox, C.D, Martinac, B, Groot, B, Walz, T. | | Deposit date: | 2020-02-27 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Visualization of the mechanosensitive ion channel MscS under membrane tension.
Nature, 590, 2021
|
|
6VYL
 
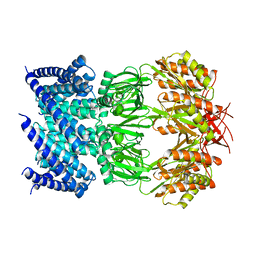 | | Cryo-EM structure of mechanosensitive channel MscS in PC-10 nanodiscs | | Descriptor: | Mechanosensitive channel MscS | | Authors: | Zhang, Y, Daday, C, Gu, R, Cox, C.D, Martinac, B, Groot, B, Walz, T. | | Deposit date: | 2020-02-27 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Visualization of the mechanosensitive ion channel MscS under membrane tension.
Nature, 590, 2021
|
|
6VYK
 
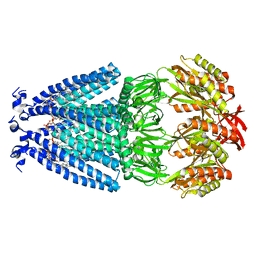 | | Cryo-EM structure of mechanosensitive channel MscS in PC-18:1 nanodiscs | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mechanosensitive channel MscS | | Authors: | Zhang, Y, Daday, C, Gu, R, Cox, C.D, Martinac, B, Groot, B, Walz, T. | | Deposit date: | 2020-02-27 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualization of the mechanosensitive ion channel MscS under membrane tension.
Nature, 590, 2021
|
|
4ZO5
 
 | | PDE10 complexed with 4-isopropoxy-2-(2-(3-(4-methoxyphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl)ethyl)isoindoline-1,3-dione | | Descriptor: | 2-{2-[3-(4-methoxyphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl]ethyl}-4-(propan-2-yloxy)-1H-isoindole-1,3(2H)-dione, 3-(1-hydroxy-2-methylpropan-2-yl)-5-phenyl-3,5-dihydro-1H-imidazo[4,5-c][1,8]naphthyridine-2,4-dione, MAGNESIUM ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-05-06 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of [(11)C]MK-8193 as a PET tracer to measure target engagement of phosphodiesterase 10A (PDE10A) inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3CJO
 
 | | Crystal structure of KSP in complex with inhibitor 30 | | Descriptor: | (2S)-4-(2,5-difluorophenyl)-N-[(3R,4S)-3-fluoro-1-methylpiperidin-4-yl]-2-(hydroxymethyl)-N-methyl-2-phenyl-2,5-dihydro-1H-pyrrole-1-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2008-03-13 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. 9. Discovery of (2S)-4-(2,5-difluorophenyl)-n-[(3R,4S)-3-fluoro-1-methylpiperidin-4-yl]-2-(hydroxymethyl)-N-methyl-2-phenyl-2,5-dihydro-1H-pyrrole-1-carboxamide (MK-0731) for the treatment of taxane-refractory cancer.
J.Med.Chem., 51, 2008
|
|
2G1Q
 
 | | crystal structure of KSP in complex with inhibitor 9h | | Descriptor: | (5S)-5-(3-AMINOPROPYL)-3-(2,5-DIFLUOROPHENYL)-N-ETHYL-5-PHENYL-4,5-DIHYDRO-1H-PYRAZOLE-1-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2006-02-14 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 4: Structure-based design of 5-alkylamino-3,5-diaryl-4,5-dihydropyrazoles as potent, water-soluble inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8XRY
 
 | |
8XS5
 
 | |
8XVX
 
 | |
8XW2
 
 | |
8XVY
 
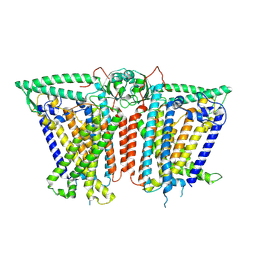 | |
8XW3
 
 | |
8XAJ
 
 | | Cryo-EM structure of OSCA1.2-liposome-inside-in open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 2024
|
|
8XS4
 
 | |
8XNG
 
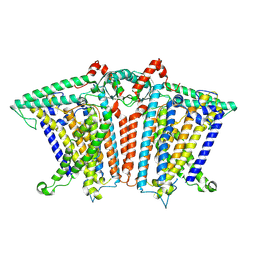 | |
8XVZ
 
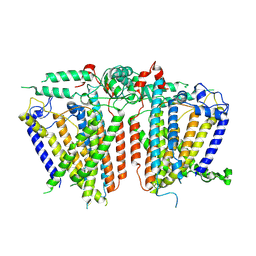 | |
8XS0
 
 | |
8XW0
 
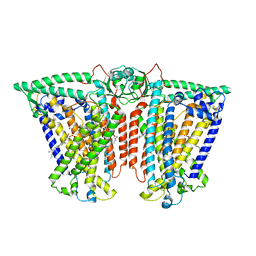 | | Cryo-EM structure of OSCA3.1-GDN state | | Descriptor: | CSC1-like protein ERD4, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 2024
|
|
8XW1
 
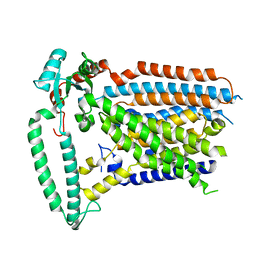 | | Cryo-EM structure of OSCA1.2-V335W-DDM state | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.49 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 2024
|
|
8XW4
 
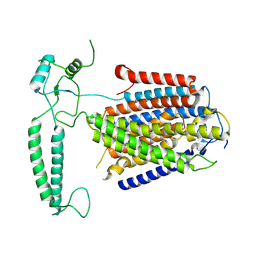 | |
8DI4
 
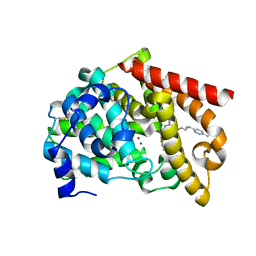 | | Discovery of MK-8189, a highly potent and selective PDE10A inhibitor for the treatment of schizophrenia | | Descriptor: | 2-methyl-6-{[(1S,2S)-2-(5-methylpyridin-2-yl)cyclopropyl]methoxy}-N-[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]pyrimidin-4-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Hayes, R.P, Yan, Y. | | Deposit date: | 2022-06-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.016 Å) | | Cite: | Discovery of MK-8189, a Highly Potent and Selective PDE10A Inhibitor for the Treatment of Schizophrenia.
J.Med.Chem., 66, 2023
|
|
7RN5
 
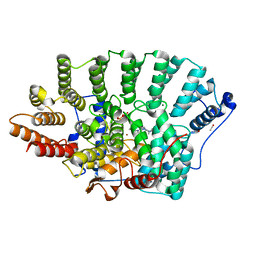 | |
7RNI
 
 | |
