6VO1
 
 | | BG505 SOSIP.v5.2 in complex with rhesus macaque Fab RM20J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2020-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
6VLR
 
 | | BG505 SOSIP.v5.2 in complex with rhesus macaque Fab RM20E1 and PGT122 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIPv5.2 gp41, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2020-01-24 | | Release date: | 2020-06-24 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
6VN0
 
 | | BG505 SOSIP.v4.1 in complex with rhesus macaque Fab RM20F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Cottrell, C.A, Shin, M, Ward, A.B. | | Deposit date: | 2020-01-29 | | Release date: | 2020-06-24 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
7LG6
 
 | | BG505 SOSIP.v5.2 in complex with VRC40.01 and RM19R Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Cottrell, C.A, Torres, J.L, Wu, N.R, Ward, A.B. | | Deposit date: | 2021-01-19 | | Release date: | 2021-09-15 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis of glycan276-dependent recognition by HIV-1 broadly neutralizing antibodies.
Cell Rep, 37, 2021
|
|
7RSN
 
 | | AMC018 SOSIP.v4.2 in complex with PGV04 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC018 gp120, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The Glycan Hole Area of HIV-1 Envelope Trimers Contributes Prominently to the Induction of Autologous Neutralization.
J.Virol., 96, 2022
|
|
6MB3
 
 | |
6MHG
 
 | |
6VKN
 
 | | BG505 SOSIP.v5.2.N241.N289 in complex with rhesus macaque Fab RM19R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-08-12 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Targeting HIV Env immunogens to B cell follicles in nonhuman primates through immune complex or protein nanoparticle formulations.
NPJ Vaccines, 5, 2020
|
|
6NC3
 
 | | AMC011 v4.2 SOSIP Env trimer in complex with fusion peptide targeting antibody VRC34 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env AMC011 v4.2 SOSIP gp120, ... | | Authors: | Cottrell, C.A, Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2018-12-10 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies.
Cell Host Microbe, 25, 2019
|
|
6NC2
 
 | | AMC011 v4.2 SOSIP Env trimer in complex with fusion peptide targeting antibody ACS202 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC011 v4.2 SOSIP gp120, ... | | Authors: | Cottrell, C.A, Ozorowski, G, Yuan, M, Copps, J, Wilson, I.A, Ward, A.B. | | Deposit date: | 2018-12-10 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies.
Cell Host Microbe, 25, 2019
|
|
6NIJ
 
 | | PGT145 Fab in complex with full length AMC011 HIV-1 Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC011 Glycoprotein 120, ... | | Authors: | Cottrell, C.A, Torrents de la Pena, A, Rantalainen, K, Torres, J.L, Ward, A.B. | | Deposit date: | 2018-12-29 | | Release date: | 2019-07-31 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics.
Plos Pathog., 15, 2019
|
|
6OZC
 
 | | BG505 SOSIP.664 with 2G12 Fab2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 Fab Heavy chain, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Networks of HIV-1 Envelope Glycans Maintain Antibody Epitopes in the Face of Glycan Additions and Deletions.
Structure, 28, 2020
|
|
6VO3
 
 | | AMC009 SOSIP.v4.2 in complex with PGV04 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC009 SOSIP.v4.2 envelope glycoprotein gp120, ... | | Authors: | Cottrell, C.A, de Val, N, Ward, A.B. | | Deposit date: | 2020-01-29 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Neutralizing Antibody Responses Induced by HIV-1 Envelope Glycoprotein SOSIP Trimers Derived from Elite Neutralizers.
J.Virol., 94, 2020
|
|
7RSO
 
 | | AMC016 SOSIP.v4.2 in complex with PGV04 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC016 gp120, ... | | Authors: | Bader, D.L.V, Cottrell, C.A, Ward, A.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The Glycan Hole Area of HIV-1 Envelope Trimers Contributes Prominently to the Induction of Autologous Neutralization.
J.Virol., 96, 2022
|
|
6DJB
 
 | | Structure of human Volume Regulated Anion Channel composed of SWELL1 (LRRC8A) | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Kefauver, J.M, Saotome, K, Pallesen, J, Cottrell, C.A, Ward, A.B, Patapoutian, A. | | Deposit date: | 2018-05-24 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of the human volume regulated anion channel.
Elife, 7, 2018
|
|
6X96
 
 | |
6X97
 
 | |
6X98
 
 | |
5I08
 
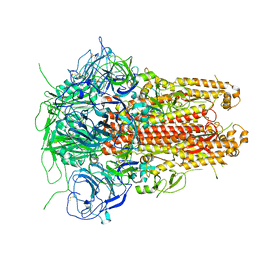 | | Prefusion structure of a human coronavirus spike protein | | Descriptor: | Spike glycoprotein,Foldon chimera | | Authors: | Kirchdoerfer, R.N, Cottrell, C.A, Wang, N, Pallesen, J, Yassine, H.M, Turner, H.L, Corbett, K.S, Graham, B.S, McLellan, J.S, Ward, A.B. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pre-fusion structure of a human coronavirus spike protein.
Nature, 531, 2016
|
|
5KEM
 
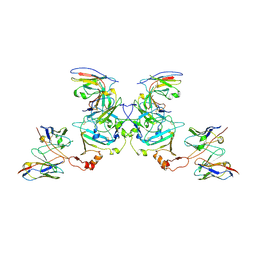 | | EBOV sGP in complex with variable Fab domains of IgGs c13C6 and BDBV91 | | Descriptor: | BDBV91 variable Fab domain heavy chain, BDBV91 variable Fab domain light chain, Ebola secreted glycoprotein, ... | | Authors: | Pallesen, J, Murin, C.D, de Val, N, Cottrell, C.A, Hastie, K.M, Turner, H.L, Fusco, M.L, Flyak, A.I, Zeitlin, L, Crowe Jr, J.E, Andersen, K.G, Saphire, E.O, Ward, A.B. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of Ebola virus GP and sGP in complex with therapeutic antibodies.
Nat Microbiol, 1, 2016
|
|
5KEL
 
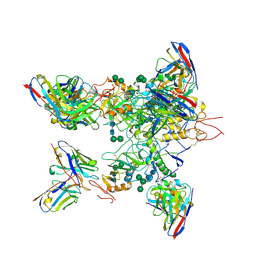 | | EBOV GP in complex with variable Fab domains of IgGs c2G4 and c13C6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ebola surface glycoprotein, ... | | Authors: | Pallesen, J, Murin, C.D, de Val, N, Cottrell, C.A, Hastie, K.M, Turner, H.L, Fusco, M.L, Flyak, A.I, Zeitlin, L, Crowe Jr, J.E, Andersen, K.G, Saphire, E.O, Ward, A.B. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of Ebola virus GP and sGP in complex with therapeutic antibodies.
Nat Microbiol, 1, 2016
|
|
5KEN
 
 | | EBOV GP in complex with variable Fab domains of IgGs c4G7 and c13C6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ebola surface glycoprotein, ... | | Authors: | Pallesen, J, Murin, C.D, de Val, N, Cottrell, C.A, Hastie, K.M, Turner, H.L, Fusco, M.L, Flyak, A.I, Zeitlin, L, Crowe Jr, J.E, Andersen, K.G, Saphire, E.O, Ward, A.B. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of Ebola virus GP and sGP in complex with therapeutic antibodies.
Nat Microbiol, 1, 2016
|
|
6CRV
 
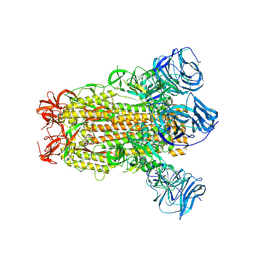 | | SARS Spike Glycoprotein, Stabilized variant, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRZ
 
 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CS0
 
 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, one S1 CTD in an upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
