3SID
 
 | | Crystal structure of oxidized Symerythrin from Cyanophora paradoxa, azide adduct at 50% occupancy | | Descriptor: | AZIDE ION, FE (III) ION, Symerythrin | | Authors: | Cooley, R.B, Arp, D.J, Karplus, P.A. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Symerythrin structures at atomic resolution and the origins of rubrerythrins and the ferritin-like superfamily.
J.Mol.Biol., 413, 2011
|
|
3QHB
 
 | |
3QHC
 
 | |
4ND7
 
 | | Crystal structure of apo 3-nitro-tyrosine tRNA synthetase (5B) in the closed form | | Descriptor: | BETA-MERCAPTOETHANOL, SODIUM ION, Tyrosine--tRNA ligase | | Authors: | Cooley, R.B, Driggers, C.M, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2013-10-25 | | Release date: | 2014-03-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Improved Second-Generation 3-Nitro-tyrosine tRNA Synthetases.
Biochemistry, 53, 2014
|
|
4NDA
 
 | | Crystal structure of 3-nitro-tyrosine tRNA synthetase (5B) bound to 3-nitro-tyrosine | | Descriptor: | GLYCEROL, META-NITRO-TYROSINE, SODIUM ION, ... | | Authors: | Cooley, R.B, Driggers, C.M, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2013-10-25 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Improved Second-Generation 3-Nitro-tyrosine tRNA Synthetases.
Biochemistry, 53, 2014
|
|
4ND6
 
 | | Crystal structure of apo 3-nitro-tyrosine tRNA synthetase (5B) in the open form | | Descriptor: | GLYCEROL, Tyrosine-tRNA ligase | | Authors: | Cooley, R.B, Driggers, C.M, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2013-10-25 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Improved Second-Generation 3-Nitro-tyrosine tRNA Synthetases.
Biochemistry, 53, 2014
|
|
4PBT
 
 | | Crystal structure of the M. jannaschii G2 tRNA synthetase variant bound to 4-trans-cyclooctene-amidopheylalanine (Tco-amF) | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 4-{[(1R,4E)-cyclooct-4-en-1-ylcarbonyl]amino}-L-phenylalanine, GLYCEROL, ... | | Authors: | Cooley, R.B, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gleaning Unexpected Fruits from Hard-Won Synthetases: Probing Principles of Permissivity in Non-canonical Amino Acid-tRNA Synthetases.
Chembiochem, 15, 2014
|
|
4PBS
 
 | |
4PBR
 
 | | Crystal structure of the M. jannaschii G2 tRNA synthetase variant bound to 4-(2-bromoisobutyramido)-phenylalanine (BibaF) | | Descriptor: | 4-[(2-bromo-2-methylpropanoyl)amino]-L-phenylalanine, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cooley, R.B, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gleaning Unexpected Fruits from Hard-Won Synthetases: Probing Principles of Permissivity in Non-canonical Amino Acid-tRNA Synthetases.
Chembiochem, 15, 2014
|
|
7Q16
 
 | | Human 14-3-3 zeta fused to the BAD peptide including phosphoserine-74 | | Descriptor: | 14-3-3 protein zeta/delta,Bcl2-associated agonist of cell death | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Gushchin, I, Remeeva, A, Kovalev, K, Cooley, R.B. | | Deposit date: | 2021-10-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Crystal structure of human 14-3-3 zeta complexed with the noncanonical phosphopeptide from proapoptotic BAD.
Biochem.Biophys.Res.Commun., 583, 2021
|
|
8DQH
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to ATP and acridone after 24 hours of crystal growth | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
8DQG
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to AMPPNP and acridone | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
8DQI
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to ATP and acridone after 2- weeks of crystal growth | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
8DQJ
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (AST) bound to ATP and acridone | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
8EQ8
 
 | |
8EQH
 
 | |
6WRT
 
 | | Crystal structure of Mj 3-nitro-tyrosine tRNA synthetase (5B) C70A/S158C variant bound to 3-nitro-tyrosine | | Descriptor: | META-NITRO-TYROSINE, SODIUM ION, Tyrosine--tRNA ligase | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
6WRK
 
 | | Crystal structure of 3rd-generation Mj 3-nitro-tyrosine tRNA synthetase ("A7") bound to 3-nitro-tyrosine | | Descriptor: | GLYCEROL, META-NITRO-TYROSINE, SODIUM ION, ... | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
6WRN
 
 | | Crystal structure of Mj 3-nitro-tyrosine tRNA synthetase (5B) C70A variant bound to 3-nitro-tyrosine | | Descriptor: | META-NITRO-TYROSINE, SODIUM ION, Tyrosine--tRNA ligase | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
6WRQ
 
 | | Crystal structure of Mj 3-nitro-tyrosine tRNA synthetase (5B) S158C variant bound to 3-nitro-tyrosine | | Descriptor: | GLYCEROL, META-NITRO-TYROSINE, SODIUM ION, ... | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
4U64
 
 | | Structure of the periplasmic output domain of the Legionella pneumophila LapD ortholog CdgS9 in the apo state | | Descriptor: | Two component histidine kinase, GGDEF domain protein/EAL domain protein | | Authors: | Chatterjee, D, Cooley, R.B, Boyd, C.D, Mehl, R.A, O'Toole, G.A, Sondermann, H.S. | | Deposit date: | 2014-07-27 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP.
Elife, 3, 2014
|
|
4U65
 
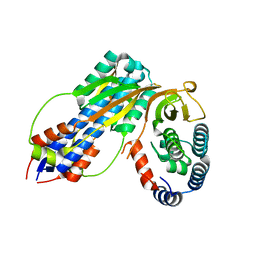 | | Structure of the periplasmic output domain of the Legionella pneumophila LapD ortholog CdgS9 in complex with Pseudomonas fluorescens LapG | | Descriptor: | CALCIUM ION, Putative cystine protease, Two component histidine kinase, ... | | Authors: | Chatterjee, D, Cooley, R.B, Boyd, C.D, Mehl, R.A, O'Toole, G.A, Sondermann, H.S. | | Deposit date: | 2014-07-27 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP.
Elife, 3, 2014
|
|
6PWK
 
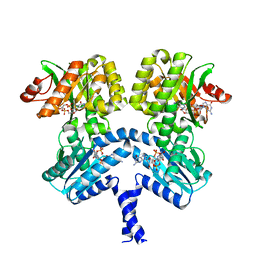 | | Vibrio cholerae LapD S helix-GGDEF-EAL (bound to c-di-GMP) | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GGDEF and EAL domain-containing protein, MAGNESIUM ION | | Authors: | Giglio, K.M, Cooley, R.B, Sondermann, H. | | Deposit date: | 2019-07-23 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | A Conserved Regulatory Circuit Controls Large Adhesins in Vibrio cholerae.
Mbio, 10, 2019
|
|
6PWJ
 
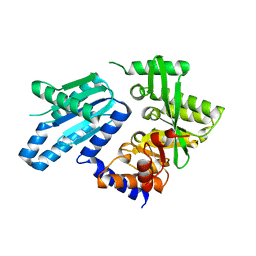 | |
4IES
 
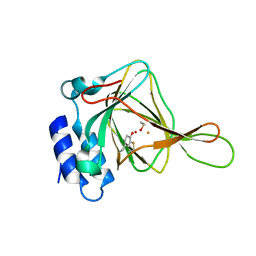 | | Cys-persulfenate bound Cysteine Dioxygenase at pH 6.2 in the presence of Cys | | Descriptor: | Cysteine dioxygenase type 1, FE (III) ION, S-HYDROPEROXYCYSTEINE | | Authors: | Driggers, C.M, Cooley, R.B, Sankaran, B, Karplus, P.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-06-26 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cysteine Dioxygenase Structures from pH4 to 9: Consistent Cys-Persulfenate Formation at Intermediate pH and a Cys-Bound Enzyme at Higher pH.
J.Mol.Biol., 425, 2013
|
|
