4FUP
 
 | |
4FUN
 
 | |
4FUM
 
 | |
4FUO
 
 | |
5CQJ
 
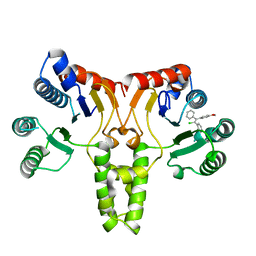 | | Crystal structure of E. coli undecaprenyl pyrophosphate synthase in complex with clomiphene | | Descriptor: | Clomifene, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Worrall, L.J, Conrady, D.G, Strynadka, N.C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Antagonism screen for inhibitors of bacterial cell wall biogenesis uncovers an inhibitor of undecaprenyl diphosphate synthase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CQB
 
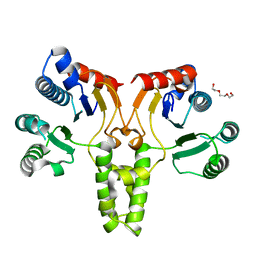 | | Crystal structure of E. coli undecaprenyl pyrophosphate synthase | | Descriptor: | Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific), TRIETHYLENE GLYCOL | | Authors: | Worrall, L.J, Conrady, D.G, Strynadka, N.C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonism screen for inhibitors of bacterial cell wall biogenesis uncovers an inhibitor of undecaprenyl diphosphate synthase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5EIQ
 
 | |
5TU8
 
 | |
5TU7
 
 | |
5TU9
 
 | |
3MXG
 
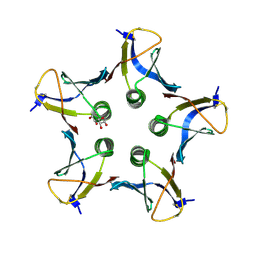 | | Structure of Shiga Toxin type 2 (Stx2) B Pentamer Mutant Q40L | | Descriptor: | D-xylose, Shiga-like toxin 2 subunit B | | Authors: | Kovall, R.A, Vander Wielen, B.D, Friedmann, D.R, Flagler, M.J. | | Deposit date: | 2010-05-07 | | Release date: | 2011-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis of differential B-pentamer stability of Shiga toxins 1 and 2.
Plos One, 5, 2010
|
|
5HX9
 
 | |
5I2B
 
 | |
5J92
 
 | |
5JC8
 
 | |
6DJ8
 
 | |
6DLK
 
 | |
6DM6
 
 | |
6NBO
 
 | |
6ONN
 
 | |
5TD3
 
 | |
5U2I
 
 | |
5UAI
 
 | |
5TP4
 
 | |
5VBF
 
 | |
