1BA4
 
 | | THE SOLUTION STRUCTURE OF AMYLOID BETA-PEPTIDE (1-40) IN A WATER-MICELLE ENVIRONMENT. IS THE MEMBRANE-SPANNING DOMAIN WHERE WE THINK IT IS? NMR, 10 STRUCTURES | | Descriptor: | AMYLOID BETA-PEPTIDE | | Authors: | Coles, M, Bicknell, W, Watson, A.A, Fairlie, D.P, Craik, D.J. | | Deposit date: | 1998-04-07 | | Release date: | 1998-06-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide(1-40) in a water-micelle environment. Is the membrane-spanning domain where we think it is?
Biochemistry, 37, 1998
|
|
1NGL
 
 | | HUMAN NEUTROPHIL GELATINASE-ASSOCIATED LIPOCALIN (HNGAL), REGULARISED AVERAGE NMR STRUCTURE | | Descriptor: | PROTEIN (NGAL) | | Authors: | Coles, M, Diercks, T, Muehlenweg, B, Bartsch, S, Zoelzer, V, Tschesche, H, Kessler, H. | | Deposit date: | 1999-02-23 | | Release date: | 1999-05-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of human neutrophil gelatinase-associated lipocalin.
J.Mol.Biol., 289, 1999
|
|
2GLW
 
 | | The solution structure of PHS018 from pyrococcus horikoshii | | Descriptor: | 92aa long hypothetical protein | | Authors: | Coles, M, Hulko, M, Truffault, V, Martin, J, Lupas, A.N. | | Deposit date: | 2006-04-05 | | Release date: | 2006-12-05 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Common evolutionary origin of swapped-hairpin and double-psi beta barrels
Structure, 14, 2006
|
|
1CZ5
 
 | | NMR STRUCTURE OF VAT-N: THE N-TERMINAL DOMAIN OF VAT (VCP-LIKE ATPASE OF THERMOPLASMA) | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Coles, M, Diercks, T, Liermann, J, Groeger, A, Rockel, B, Baumeister, W, Koretke, K, Lupas, A, Peters, J, Kessler, H. | | Deposit date: | 1999-09-01 | | Release date: | 1999-10-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of VAT-N reveals a 'missing link' in the evolution of complex enzymes from a simple betaalphabetabeta element.
Curr.Biol., 9, 1999
|
|
1CZ4
 
 | | NMR STRUCTURE OF VAT-N: THE N-TERMINAL DOMAIN OF VAT (VCP-LIKE ATPASE OF THERMOPLASMA) | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Coles, M, Diercks, T, Liermann, J, Groeger, A, Rockel, B, Baumeister, W, Koretke, K, Lupas, A, Peters, J, Kessler, H. | | Deposit date: | 1999-09-01 | | Release date: | 1999-10-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of VAT-N reveals a 'missing link' in the evolution of complex enzymes from a simple betaalphabetabeta element.
Curr.Biol., 9, 1999
|
|
2L7H
 
 | |
6TH8
 
 | |
1YFB
 
 | |
1YSF
 
 | |
2L7I
 
 | |
2LFS
 
 | | Solution structure of the chimeric Af1503 HAMP- EnvZ DHp homodimer; A219F variant | | Descriptor: | HAMP domain-containing protein, Osmolarity sensor protein EnzV chimera | | Authors: | Coles, M, Ferris, H.U, Hulko, M, Martin, J, Lupas, A.N. | | Deposit date: | 2011-07-10 | | Release date: | 2011-08-24 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | Mechanism of regulation of receptor histidine kinases.
Structure, 20, 2012
|
|
2MPW
 
 | | Solution structure of the LysM region of the E. coli Intimin periplasmic domain | | Descriptor: | Intimin | | Authors: | Coles, M, Chaubey, M, Leo, J.C, Linke, D, Schuetz, M.C, Goetz, F, Autenrieth, I.B. | | Deposit date: | 2014-06-05 | | Release date: | 2014-11-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Intimin periplasmic domain mediates dimerisation and binding to peptidoglycan.
Mol.Microbiol., 95, 2015
|
|
2LFR
 
 | | Solution structure of the chimeric Af1503 HAMP- EnvZ DHp homodimer | | Descriptor: | HAMP domain-containing protein, Osmolarity sensor protein EnzV chimera | | Authors: | Coles, M, Ferris, H.U, Hulko, M, Martin, J, Lupas, A.N. | | Deposit date: | 2011-07-10 | | Release date: | 2011-08-24 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | Mechanism of regulation of receptor histidine kinases.
Structure, 20, 2012
|
|
2LDY
 
 | |
2LLE
 
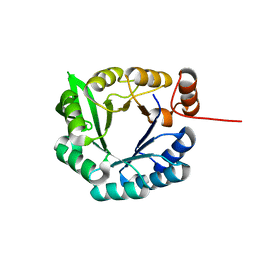 | | Computational design of an eight-stranded (beta/alpha)-barrel from fragments of different folds | | Descriptor: | Chemotaxis protein CheY, Imidazole glycerol phosphate synthase subunit HisF chimera | | Authors: | Coles, M, Truffault, V, Eisenbeis, S, Proffitt, W, Meiler, J, Hocker, B. | | Deposit date: | 2011-11-07 | | Release date: | 2012-03-21 | | Method: | SOLUTION NMR | | Cite: | Potential of fragment recombination for rational design of proteins.
J.Am.Chem.Soc., 134, 2012
|
|
2P3M
 
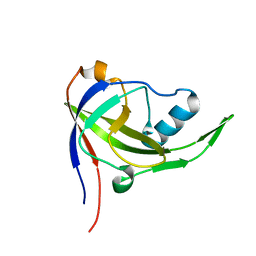 | | Solution structure of Mj0056 | | Descriptor: | Riboflavin Kinase MJ0056 | | Authors: | Coles, M, Truffault, V, Djuranovic, S, Martin, J, Lupas, A.N. | | Deposit date: | 2007-03-09 | | Release date: | 2007-10-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A CTP-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
7NY0
 
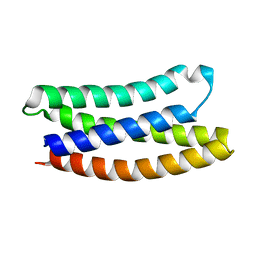 | |
8B1X
 
 | | Solution NMR structure of the single alpha helix peptide (P3-7)2 | | Descriptor: | P3-7_2 | | Authors: | Escobedo, A, Coles, M, Diercks, T, Garcia, J, Millet, O, Salvatella, X. | | Deposit date: | 2022-09-12 | | Release date: | 2023-01-25 | | Method: | SOLUTION NMR | | Cite: | A glutamine-based single alpha-helix scaffold to target globular proteins.
Nat Commun, 13, 2022
|
|
6Y06
 
 | |
6Y07
 
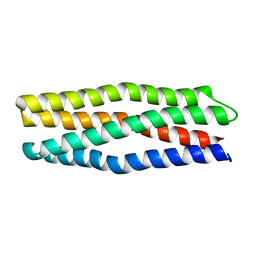 | |
1SVJ
 
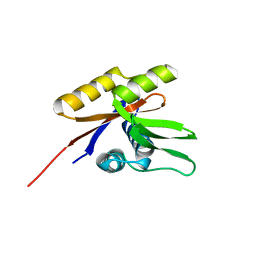 | | The solution structure of the nucleotide binding domain of KdpB | | Descriptor: | Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2004-03-29 | | Release date: | 2004-09-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Inter-domain motions of the N-domain of the KdpFABC complex, a P-type ATPase, are not driven by ATP-induced conformational changes.
J.Mol.Biol., 342, 2004
|
|
1U7Q
 
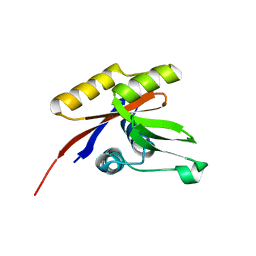 | | THE SOLUTION STRUCTURE OF THE NUCLEOTIDE BINDING DOMAIN OF KDPB | | Descriptor: | Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2004-08-04 | | Release date: | 2004-09-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Inter-domain motions of the N-domain of the KdpFABC complex, a P-type ATPase, are not driven by ATP-induced conformational changes.
J.Mol.Biol., 342, 2004
|
|
6QF8
 
 | |
6QFP
 
 | |
6QH2
 
 | |
