1DIC
 
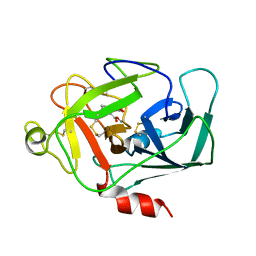 | | STRUCTURE OF 3,4-DICHLOROISOCOUMARIN-INHIBITED FACTOR D | | Descriptor: | 3,4-DICHLOROISOCOUMARIN, FACTOR D, OXYGEN ATOM | | Authors: | Cole, L.B, Kilpatrick, J.M, Chu, N, Babu, Y.S. | | Deposit date: | 1998-07-08 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of 3,4-dichloroisocoumarin-inhibited factor D.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1DFP
 
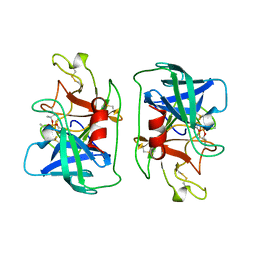 | | FACTOR D INHIBITED BY DIISOPROPYL FLUOROPHOSPHATE | | Descriptor: | DIISOPROPYL PHOSPHONATE, FACTOR D | | Authors: | Cole, L.B, Chu, N, Kilpatrick, J.M, Volanakis, J.E, Narayana, S.V.L, Babu, Y.S. | | Deposit date: | 1997-02-18 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of diisopropyl fluorophosphate-inhibited factor D.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
2DRI
 
 | |
1DRJ
 
 | |
1TPT
 
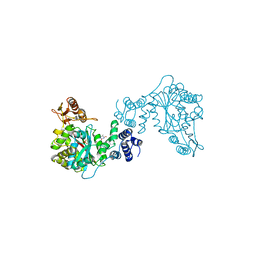 | | THREE-DIMENSIONAL STRUCTURE OF THYMIDINE PHOSPHORYLASE FROM ESCHERICHIA COLI AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | SULFATE ION, THYMIDINE PHOSPHORYLASE, THYMINE | | Authors: | Walter, M.R, Cook, W.J, Cole, L.B, Short, S.A, Koszalka, G.W, Krenitsky, T.A, Ealick, S.E. | | Deposit date: | 1990-06-14 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of thymidine phosphorylase from Escherichia coli at 2.8 A resolution.
J.Biol.Chem., 265, 1990
|
|
1BA2
 
 | |
1DBP
 
 | |
1DRK
 
 | |
1URP
 
 | |
1AZY
 
 | | STRUCTURAL AND THEORETICAL STUDIES SUGGEST DOMAIN MOVEMENT PRODUCES AN ACTIVE CONFORMATION OF THYMIDINE PHOSPHORYLASE | | Descriptor: | THYMIDINE PHOSPHORYLASE | | Authors: | Pugmire, M.J, Cook, W.J, Jasanoff, A, Walter, M.R, Ealick, S.E. | | Deposit date: | 1997-11-24 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and theoretical studies suggest domain movement produces an active conformation of thymidine phosphorylase.
J.Mol.Biol., 281, 1998
|
|
3GBP
 
 | |
1GCA
 
 | |
1GCG
 
 | |
2TPT
 
 | | STRUCTURAL AND THEORETICAL STUDIES SUGGEST DOMAIN MOVEMENT PRODUCES AN ACTIVE CONFORMATION OF THYMIDINE PHOSPHORYLASE | | Descriptor: | SULFATE ION, THYMIDINE PHOSPHORYLASE | | Authors: | Pugmire, M.J, Cook, W.J, Jasanoff, A, Walter, M.R, Ealick, S.E. | | Deposit date: | 1997-11-24 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and theoretical studies suggest domain movement produces an active conformation of thymidine phosphorylase.
J.Mol.Biol., 281, 1998
|
|
