3RON
 
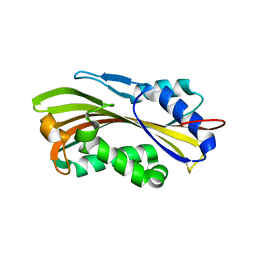 | | Crystal Structure and Hemolytic Activity of the Cyt1Aa Toxin from Bacillus thuringiensis subsp. israelensis | | Descriptor: | Type-1Aa cytolytic delta-endotoxin | | Authors: | Cohen, S, Albeck, S, Ben-Dov, E, Cahan, R, Firer, M, Zaritsky, A, Dym, O, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-04-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Cyt1Aa Toxin: Crystal Structure Reveals Implications for Its Membrane-Perforating Function.
J.Mol.Biol., 413, 2011
|
|
8E8Q
 
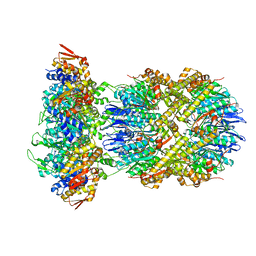 | | Cryo-EM structure of substrate-free DNClpX.ClpP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ghanbarpour, A, Cohen, S, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-08-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structure of substrate-free DNClpX.ClpP
Nat Commun, 2023
|
|
8E7V
 
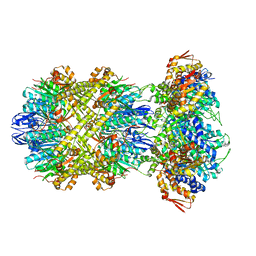 | | Cryo-EM structure of substrate-free DNClpX.ClpP from singly capped particles | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ghanbarpour, A, Cohen, S, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-08-24 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of substrate-free DNClpX.ClpP
Nat Commun, 2023
|
|
2RCI
 
 | |
6XXO
 
 | |
6XXP
 
 | |
6XXN
 
 | | Crystal structure of NB7, a nanobody targeting prostate specific membrane antigen | | Descriptor: | NB_7_a,b,c,f, NB_7_g, NB_7_h, ... | | Authors: | Shahar, A, Rosenfeld, L, Papo, N. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Nanobodies Targeting Prostate-Specific Membrane Antigen for the Imaging and Therapy of Prostate Cancer.
J.Med.Chem., 63, 2020
|
|
7SEG
 
 | | Crystal structure of the complex of CD16A bound by an anti-CD16A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Kiefer, J.R, Wallweber, H.A, Polson, A.G. | | Deposit date: | 2021-09-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | A BCMA/CD16A bispecific innate cell engager for the treatment of multiple myeloma.
Leukemia, 36, 2022
|
|
5LGY
 
 | |
4XMK
 
 | |
4XML
 
 | | Crystal structure of Fab of HIV-1 gp120 V3-specific human monoclonal antibody 2424 | | Descriptor: | Heavy chain of HIV-1 gp120 V3-specific human monoclonal antibody 2424, Light chain of HIV-1 gp120 V3-specific human monoclonal antibody 2424 | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2015-01-14 | | Release date: | 2015-07-08 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Functional and Structural Characterization of Human V3-Specific Monoclonal Antibody 2424 with Neutralizing Activity against HIV-1 JRFL.
J.Virol., 89, 2015
|
|
6C3L
 
 | | Crystal structure of BCL6 BTB domain with compound 15f | | Descriptor: | B-cell lymphoma 6 protein, N-[2-(1H-indol-3-yl)ethyl]-N'-{3-[(4-methylpiperazin-1-yl)methyl]-1-[2-(morpholin-4-yl)-2-oxoethyl]-1H-indol-6-yl}thiourea | | Authors: | Linhares, B, Cheng, H, Cierpicki, T, Xue, F. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.46092153 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
6C3N
 
 | | Crystal structure of BCL6 BTB domain in complex with compound 7CC5 | | Descriptor: | B-cell lymphoma 6 protein, N-(2-phenylethyl)-N'-pyridin-3-ylthiourea | | Authors: | Linhares, B, Cheng, H, Xue, F, Cierpicki, T. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53170586 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
6CQ1
 
 | | BCL6 BTB domain in complex with 15a | | Descriptor: | 2-{6-({[2-(1H-indol-3-yl)ethyl]carbamothioyl}amino)-3-[(4-methylpiperazin-1-yl)methyl]-1H-indol-1-yl}-N-(propan-2-yl)acetamide, B-cell lymphoma 6 protein | | Authors: | Linhares, B.M, Cheng, H, Xue, F, Cierpicki, T. | | Deposit date: | 2018-03-14 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69921041 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
8E91
 
 | |
5TH4
 
 | |
5THJ
 
 | | Crystal Structure of 2-hydroxycyclohepta-2,4,6-trien-1-one bound to human carbonic anhydrase 2 | | Descriptor: | 2-HYDROXYCYCLOHEPTA-2,4,6-TRIEN-1-ONE, CITRIC ACID, Carbonic anhydrase 2, ... | | Authors: | Dick, B, Cohen, S. | | Deposit date: | 2016-09-29 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effect of donor atom identity on metal-binding pharmacophore coordination.
J. Biol. Inorg. Chem., 22, 2017
|
|
5TI0
 
 | | Crystal Structure of 2-Hydroxycyclohepta-2,4,6-triene-1-thione bound to human carbonic anhydrase 2 L198G | | Descriptor: | 2-hydroxycyclohepta-2,4,6-triene-1-thione, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Dick, B, Cohen, S. | | Deposit date: | 2016-09-30 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Effect of donor atom identity on metal-binding pharmacophore coordination.
J. Biol. Inorg. Chem., 22, 2017
|
|
5THI
 
 | | Crystal Structure of 2-hydroxycyclohepta-2,4,6-trien-1-one bound to human carbonic anhydrase 2 L198G | | Descriptor: | 2-HYDROXYCYCLOHEPTA-2,4,6-TRIEN-1-ONE, CITRIC ACID, Carbonic anhydrase 2, ... | | Authors: | Dick, B, Cohen, S. | | Deposit date: | 2016-09-29 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effect of donor atom identity on metal-binding pharmacophore coordination.
J. Biol. Inorg. Chem., 22, 2017
|
|
5THN
 
 | | Crystal Structure of 2-Hydroxycyclohepta-2,4,6-triene-1-thione bound to human carbonic anhydrase 2 | | Descriptor: | 2-hydroxycyclohepta-2,4,6-triene-1-thione, CITRIC ACID, Carbonic anhydrase 2, ... | | Authors: | Dick, B, Cohen, S. | | Deposit date: | 2016-09-29 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Effect of donor atom identity on metal-binding pharmacophore coordination.
J. Biol. Inorg. Chem., 22, 2017
|
|
5BUA
 
 | |
4UAO
 
 | |
