3RQW
 
 | | Crystal structure of acetylcholine bound to a prokaryotic pentameric ligand-gated ion channel, ELIC | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETYLCHOLINE, ELIC Pentameric Ligand Gated Ion Channel from Erwinia Chrysanthemi, ... | | Authors: | Pan, J.J, Chen, Q, Yoshida, K, Cohen, A, Kong, X.P, Xu, Y, Tang, P. | | Deposit date: | 2011-04-28 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine.
Nat Commun, 3, 2012
|
|
3RQU
 
 | | Crystal structure of a prokaryotic pentameric ligand-gated ion channel, ELIC | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ELIC Pentameric Ligand Gated Ion Channel from Erwinia Chrysanthemi, GLYCEROL | | Authors: | Pan, J.J, Chen, Q, Yoshida, K, Cohen, A, Kong, X.P, Xu, Y, Tang, P. | | Deposit date: | 2011-04-28 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine.
Nat Commun, 3, 2012
|
|
6EM0
 
 | | Crystal Structure of 2-hydroxybiphenyl 3-monooxygenase M321A from Pseudomonas azelaica | | Descriptor: | 2-hydroxybiphenyl-3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Deri, B, Bregman-Cohen, A, Pazy Benhar, Y, Fishman, A. | | Deposit date: | 2017-10-01 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Altering 2-Hydroxybiphenyl 3-Monooxygenase Regioselectivity by Protein Engineering for the Production of a New Antioxidant.
Chembiochem, 19, 2018
|
|
4Z2R
 
 | |
4Z2U
 
 | |
4Z2T
 
 | |
5BRT
 
 | | Crystal Structure of 2-hydroxybiphenyl 3-monooxygenase from Pseudomonas azelaica with 2-hydroxybiphenyl in the active site | | Descriptor: | 2-HYDROXYBIPHENYL, 2-hydroxybiphenyl-3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kanteev, M, Bregman-Cohen, A, Deri, B, Adir, N, Fishman, A. | | Deposit date: | 2015-06-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A crystal structure of 2-hydroxybiphenyl 3-monooxygenase with bound substrate provides insights into the enzymatic mechanism.
Biochim.Biophys.Acta, 1854, 2015
|
|
4F8H
 
 | | X-ray Structure of the Anesthetic Ketamine Bound to the GLIC Pentameric Ligand-gated Ion Channel | | Descriptor: | (R)-ketamine, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Proton-gated ion channel, ... | | Authors: | Pan, J.J, Chen, Q, Willenbring, D, Kong, X.P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2012-05-17 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of the Pentameric Ligand-Gated Ion Channel GLIC Bound with Anesthetic Ketamine.
Structure, 20, 2012
|
|
4ZAR
 
 | | Crystal Structure of Proteinase K from Engyodontium albuminhibited by METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE at 1.15 A resolution | | Descriptor: | CALCIUM ION, METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE, bound form, ... | | Authors: | Sawaya, M.R, Cascio, D, Collazo, M, Bond, C, Cohen, A, DeNicola, A, Eden, K, Jain, K, Leung, C, Lubock, N, McCormick, J, Rosinski, J, Spiegelman, L, Athar, Y, Tibrewal, N, Winter, J, Solomon, S. | | Deposit date: | 2015-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of Proteinase K from Engyodontium album inhibited by METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE at 1.15 A resolution
to be published
|
|
7TIH
 
 | | Structure of oxidized bovine cytochrome c oxidase with reduced metal centers induced by synchrotron X-ray exposure | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ishigami, I, Rousseau, D.L, Yeh, S.-R, Russi, S, Cohen, A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Temperature-dependent structural transition following X-ray-induced metal center reduction in oxidized cytochrome c oxidase.
J.Biol.Chem., 298, 2022
|
|
7TII
 
 | | Annealed structure of oxidized bovine cytochrome c oxidase with reduced metal centers induced by synchrotron X-ray exposure | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ishigami, I, Rousseau, D.L, Yeh, S.-R, Russi, S, Cohen, A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Temperature-dependent structural transition following X-ray-induced metal center reduction in oxidized cytochrome c oxidase.
J.Biol.Chem., 298, 2022
|
|
5SXU
 
 | | X-ray structure of 2-bromoethanol bound to a pentameric ligand gated ion channel (ELIC) in a desensitized state | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-BROMOETHANOL, 3-AMINOPROPANE, ... | | Authors: | Chen, Q, Kinde, M, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2016-08-10 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Alcohol Inhibition of the Pentameric Ligand-Gated Ion Channel ELIC.
Structure, 25, 2017
|
|
5SXV
 
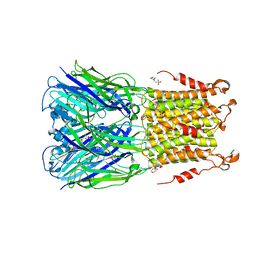 | | X-ray structure of 2-bromoethanol bound to a pentameric ligand gated ion channel (ELIC) in a resting state | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-BROMOETHANOL, Cys-loop ligand-gated ion channel | | Authors: | Chen, Q, Kinde, M, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2016-08-10 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Alcohol Inhibition of the Pentameric Ligand-Gated Ion Channel ELIC.
Structure, 25, 2017
|
|
4PNJ
 
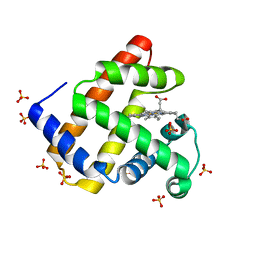 | | Recombinant Sperm Whale P6 Myoglobin Solved with Single Pulse Free Electron Laser Data | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Cohen, A, Gonzalez, A, Lam, W, Lyubimov, A, Sauter, N, Tsai, Y, Uervirojnangkoorn, M, Brunger, A, Soltis, M. | | Deposit date: | 2014-05-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Goniometer-based femtosecond crystallography with X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6APF
 
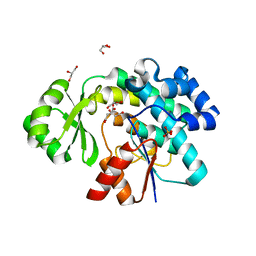 | | Trans-acting transferase from Disorazole synthase complexed with Citrate. | | Descriptor: | CITRIC ACID, DisD protein, GLYCEROL, ... | | Authors: | Mathews, I.I, Lyubimov, A, Soltis, M, Khosla, C, Cohen, A, Robbins, T. | | Deposit date: | 2017-08-17 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Biochemistry, 56, 2017
|
|
5EJX
 
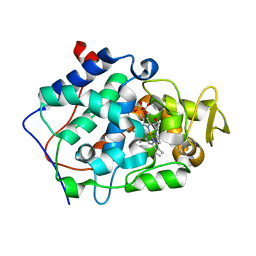 | | X-ray Free Electron Laser Structure of Cytochrome C Peroxidase | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PHOSPHATE ION, ... | | Authors: | Doukov, T, Soltis, S.M, Baxter, E.L, Cohen, A, Song, J, McPhillips, S, Poulos, T.L, Meharenna, Y.T, Chreifi, G. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the pristine peroxidase ferryl center and its relevance to proton-coupled electron transfer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EJT
 
 | | Thermally annealed ferryl Cytochrome C Peroxidase crystal structure | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PHOSPHATE ION, ... | | Authors: | Doukov, T, Soltis, S.M, Baxter, E.L, Cohen, A, Song, J, McPhillips, S, Poulos, T.L, Meharenna, Y.T, Chreifi, G. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the pristine peroxidase ferryl center and its relevance to proton-coupled electron transfer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5OLW
 
 | |
5OLY
 
 | |
7URY
 
 | | Tetradecameric hub domain of CaMKII beta | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit beta, D-MALATE | | Authors: | Ozden, C, Saha, S, Samkutty, A, Stratton, M.M, Garman, S.C, Perry, S.L. | | Deposit date: | 2022-04-22 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Polymer-based microfluidic device for on-chip counter-diffusive crystallization and in situ X-ray crystallography at room temperature.
Lab Chip, 23, 2023
|
|
1C3P
 
 | |
1C3S
 
 | |
1C3R
 
 | |
4Z90
 
 | | ELIC bound with the anesthetic isoflurane in the resting state | | Descriptor: | (2R)-2-chloro-2-(difluoromethoxy)-1,1,1-trifluoroethane, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Chen, Q, Kinde, M, Arjunan, P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Direct Pore Binding as a Mechanism for Isoflurane Inhibition of the Pentameric Ligand-gated Ion Channel ELIC.
Sci Rep, 5, 2015
|
|
4Z91
 
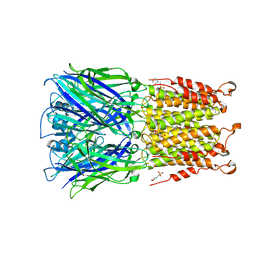 | | ELIC cocrystallized with isofluorane in a desensitized state | | Descriptor: | (2R)-2-chloro-2-(difluoromethoxy)-1,1,1-trifluoroethane, 1.7.6 3-bromanylpropan-1-amine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Chen, Q, Kinde, M.N, Arjunan, P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3915 Å) | | Cite: | Direct Pore Binding as a Mechanism for Isoflurane Inhibition of the Pentameric Ligand-gated Ion Channel ELIC.
Sci Rep, 5, 2015
|
|
