19HC
 
 | | NINE-HAEM CYTOCHROME C FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 | | Descriptor: | ACETATE ION, PROTEIN (NINE-HAEM CYTOCHROME C), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Matias, P.M, Coelho, R, Pereira, I.A.C, Coelho, A.V, Thompson, A.W, Sieker, L, Gall, J.L, Carrondo, M.A. | | Deposit date: | 1998-12-01 | | Release date: | 1999-12-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The primary and three-dimensional structures of a nine-haem cytochrome c from Desulfovibrio desulfuricans ATCC 27774 reveal a new member of the Hmc family.
Structure Fold.Des., 7, 1999
|
|
2WPN
 
 | | Structure of the oxidised, as-isolated NiFeSe hydrogenase from D. vulgaris Hildenborough | | Descriptor: | 3-[DODECYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, BIS-(MU-2-OXO),[(MU-3--SULFIDO)-BIS(MU-2--SULFIDO)-TRIS(CYS-S)-TRI-IRON] (AQUA)(GLU-O)IRON(II), CARBONMONOXIDE-(DICYANO) IRON, ... | | Authors: | Marques, M.C, Coelho, R, De Lacey, A.L, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2009-08-07 | | Release date: | 2010-01-12 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The three-dimensional structure of [NiFeSe] hydrogenase from Desulfovibrio vulgaris Hildenborough: a hydrogenase without a bridging ligand in the active site in its oxidised, "as-isolated" state.
J.Mol.Biol., 396, 2010
|
|
2VKR
 
 | | 3Fe-4S, 4Fe-4S plus Zn Acidianus ambivalens ferredoxin | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ZINC ION, ... | | Authors: | Frazao, C, Aragao, D, Coelho, R, Leal, S.S, Gomes, C.M, Teixeira, M, Carrondo, M.A. | | Deposit date: | 2007-12-23 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystallographic analysis of the intact metal centres [3Fe-4S](1+/0) and [4Fe-4S](2+/1+) in a Zn(2+) -containing ferredoxin.
FEBS Lett., 582, 2008
|
|
6QPA
 
 | | Halothiobacillus neapolitanus sulfur oxygenase reductase | | Descriptor: | FE (III) ION, SULFATE ION, Sulfur oxygenase/reductase | | Authors: | Frazao, C, Klezin, A, Poell, U, Veith, A, Ruehl, P, Coelho, R. | | Deposit date: | 2019-02-13 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multiple sulfane modifications in active-site cysteine thiols of two sulfur oxygenase reductases and analysis of substrate/product channels
To Be Published
|
|
3ZE9
 
 | | 3D structure of the NiFeSe hydrogenase from D. vulgaris Hildenborough in the oxidized as-isolated state at 1.33 Angstroms | | Descriptor: | BIS-(MU-2-OXO),[(MU-3--SULFIDO)-BIS(MU-2--SULFIDO)-TRIS(CYS-S)-TRI-IRON] (AQUA)(GLU-O)IRON(II), CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 38, 2013
|
|
3ZE8
 
 | | 3D structure of the Ni-Fe-Se hydrogenase from D. vulgaris Hildenborough in the reduced state at 1.95 Angstroms | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE (II) ION, ... | | Authors: | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 38, 2013
|
|
3ZE6
 
 | | 3D structure of the Ni-Fe-Se hydrogenase from D. vulgaris Hildenborough in the as-isolated oxidized state at 1.50 Angstroms | | Descriptor: | 3-[DODECYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, BIS-(MU-2-OXO),[(MU-3--SULFIDO)-BIS(MU-2--SULFIDO)-TRIS(CYS-S)-TRI-IRON] (AQUA)(GLU-O)IRON(II), CARBONMONOXIDE-(DICYANO) IRON, ... | | Authors: | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 2013
|
|
3ZE7
 
 | | 3D structure of the Ni-Fe-Se hydrogenase from D. vulgaris Hildenborough in the reduced state at 1.95 Angstroms | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, HYDROSULFURIC ACID, ... | | Authors: | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 38, 2013
|
|
3ZEA
 
 | | 3D structure of the NiFeSe hydrogenase from D. vulgaris Hildenborough in the reduced state at 1.82 Angstroms | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, HYDROSULFURIC ACID, ... | | Authors: | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 38, 2013
|
|
1VZM
 
 | | OSTEOCALCIN FROM FISH ARGYROSOMUS REGIUS | | Descriptor: | MAGNESIUM ION, OSTEOCALCIN | | Authors: | Frazao, C, Simes, D.C, Coelho, R, Alves, D, Williamson, M.K, Price, P.A, Cancela, M.L, Carrondo, M.A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-09-10 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Evidence of a Fourth Gla Residue in Fish Osteocalcin: Biological Implications
Biochemistry, 44, 2005
|
|
1WAD
 
 | | CYTOCHROME C3 WITH 4 HEME GROUPS AND ONE CALCIUM ION | | Descriptor: | CALCIUM ION, CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Matias, P.M, Morais, J, Coelho, R, Carrondo, M.A, Wilson, K, Dauter, Z, Sieker, L. | | Deposit date: | 1996-01-10 | | Release date: | 1997-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cytochrome c3 from Desulfovibrio gigas: crystal structure at 1.8 A resolution and evidence for a specific calcium-binding site.
Protein Sci., 5, 1996
|
|
1E5D
 
 | | RUBREDOXIN OXYGEN:OXIDOREDUCTASE (ROO) FROM ANAEROBE DESULFOVIBRIO GIGAS | | Descriptor: | FLAVIN MONONUCLEOTIDE, MU-OXO-DIIRON, OXYGEN MOLECULE, ... | | Authors: | Frazao, C, Silva, G, Gomes, C.M, Matias, P, Coelho, R, Sieker, L, Macedo, S, Liu, M.Y, Oliveira, S, Teixeira, M, Xavier, A.V, Rodrigues-Pousada, C, Carrondo, M.A, Le Gall, J. | | Deposit date: | 2000-07-24 | | Release date: | 2000-11-17 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Dioxygen Reduction Enzyme from Desulfovibrio Gigas
Nat.Struct.Biol., 7, 2000
|
|
1E29
 
 | | PSII associated cytochrome C549 from Synechocystis sp. | | Descriptor: | CALCIUM ION, CYTOCHROME C549, HEME C | | Authors: | Frazao, C, Enguita, F.J, Coelho, R, Sheldrick, G.M. | | Deposit date: | 2000-05-19 | | Release date: | 2001-05-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal Structure of Low-Potential Cytochrome C549 from Synechocystis Sp. Pcc 6803 at 1.21A Resolution
J.Biol.Inorg.Chem., 6, 2001
|
|
1E3G
 
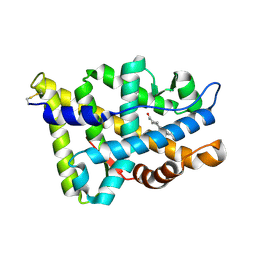 | | Human Androgen Receptor Ligand Binding in complex with the ligand metribolone (R1881) | | Descriptor: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, ANDROGEN RECEPTOR | | Authors: | Matias, P.M, Donner, P, Coelho, R, Thomaz, M, Peixoto, C, Macedo, S, Otto, N, Joschko, S, Scholz, P, Wegg, A, Basler, S, Schafer, M, Ruff, M, Egner, U, Carrondo, M.A. | | Deposit date: | 2000-06-14 | | Release date: | 2001-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations.
J. Biol. Chem., 275, 2000
|
|
1E3K
 
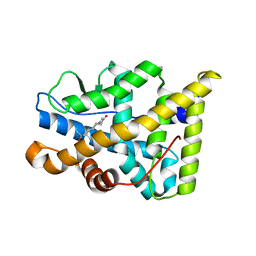 | | Human Progesteron Receptor Ligand Binding Domain in complex with the ligand metribolone (R1881) | | Descriptor: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, PROGESTERONE RECEPTOR | | Authors: | Matias, P.M, Donner, P, Coelho, R, Thomaz, M, Peixoto, C, Macedo, S, Otto, N, Joschko, S, Scholz, P, Wegg, A, Basler, S, Schafer, M, Egner, U, Carrondo, M.A. | | Deposit date: | 2000-06-19 | | Release date: | 2001-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations.
J. Biol. Chem., 275, 2000
|
|
1E3D
 
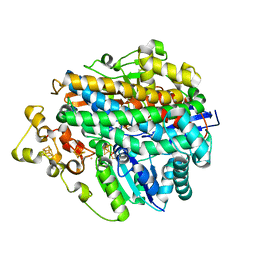 | | [NiFe] Hydrogenase from Desulfovibrio desulfuricans ATCC 27774 | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), BIS-(MU-2-OXO),[(MU-3--SULFIDO)-BIS(MU-2--SULFIDO)-TRIS(CYS-S)-TRI-IRON] (AQUA)(GLU-O)IRON(II), FE3-S4 CLUSTER, ... | | Authors: | Matias, P.M, Soares, C.M, Saraiva, L.M, Coelho, R, Morais, J, Le Gall, J, Carrondo, M.A. | | Deposit date: | 2000-06-14 | | Release date: | 2001-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | [Nife] Hydrogenase from Desulfovibrio Desulfuricans Atcc 27774: Gene Sequencing, Three-Dimensional Structure Determination and Refinement at 1.8 A and Modelling Studies of its Interaction with the Tetrahaem Cytochrome C3.
J.Biol.Inorg.Chem., 6, 2001
|
|
1GNL
 
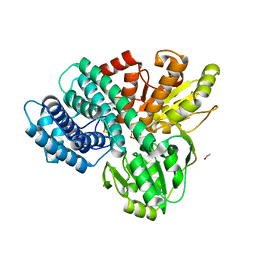 | | Hybrid Cluster Protein from Desulfovibrio desulfuricans X-ray structure at 1.25A resolution using synchrotron radiation at a wavelength of 0.933A | | Descriptor: | ACETATE ION, HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hybrid Cluster Proteins (Hcps) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at 1.25 A Resolution Using Synchrotron Radiation.
J.Biol.Inorg.Chem., 7, 2002
|
|
1GN9
 
 | | Hybrid Cluster Protein from Desulfovibrio desulfuricans ATCC 27774 X-ray structure at 2.6A resolution using synchrotron radiation at a wavelength of 1.722A | | Descriptor: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-04 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hybrid Cluster Proteins (Hcps) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at 1.25 A Resolution Using Synchrotron Radiation.
J.Biol.Inorg.Chem., 7, 2002
|
|
1GNT
 
 | | Hybrid Cluster Protein from Desulfovibrio vulgaris. X-ray structure at 1.25A resolution using synchrotron radiation. | | Descriptor: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hybrid cluster proteins (HCPs) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at 1.25 A resolution using synchrotron radiation.
J. Biol. Inorg. Chem., 7, 2002
|
|
1GS4
 
 | | Structural basis for the glucocorticoid response in a mutant human androgen receptor (ARccr) derived from an androgen-independent prostate cancer | | Descriptor: | 9ALPHA-FLUOROCORTISOL, ANDROGEN RECEPTOR, PHOSPHATE ION | | Authors: | Matias, P.M, Carrondo, M.A, Coelho, R, Thomaz, M, Zhao, X.-Y, Wegg, A, Crusius, K, Egner, U, Donner, P. | | Deposit date: | 2001-12-27 | | Release date: | 2003-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for the Glucocorticoid Response in a Mutant Human Androgen Receptor (Ar(Ccr)) Derived from an Androgen-Independent Prostate Cancer
J.Med.Chem., 45, 2002
|
|
1B5F
 
 | | NATIVE CARDOSIN A FROM CYNARA CARDUNCULUS L. | | Descriptor: | PROTEIN (CARDOSIN A), alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Frazao, C, Bento, I, Carrondo, M.A. | | Deposit date: | 1999-01-06 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of cardosin A, a glycosylated and Arg-Gly-Asp-containing aspartic proteinase from the flowers of Cynara cardunculus L.
J.Biol.Chem., 274, 1999
|
|
5MAB
 
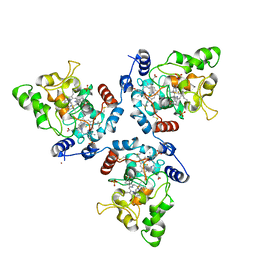 | | FoxE P3121 crystal structure of Rhodopseudomonas ferrooxidans SW2 putative iron oxidase | | Descriptor: | COPPER (II) ION, FoxE, HEME C, ... | | Authors: | Pereira, L, Saraiva, I.H, Oliveira, A.S, Soares, C, Gomes, R.O, Frazao, C. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | FoxE P3121 crystal structure of Rhodopseudomonas ferrooxidans SW2 putative iron oxidase
To be published
|
|
5MVO
 
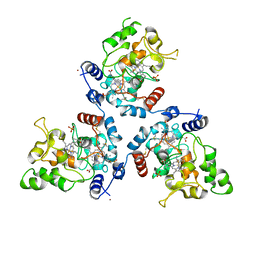 | | FoxE P43212 crystal structure of Rhodopseudomonas ferrooxidans SW2 putative iron oxidase | | Descriptor: | COPPER (II) ION, FoxE, HEME C, ... | | Authors: | Pereira, L, Saraiva, I.H, Oliveira, A.S, Soares, C, Gomes, R.O, Frazao, C. | | Deposit date: | 2017-01-17 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.668 Å) | | Cite: | Crystallization and preliminary crystallographic studies of FoxE from Rhodobacter ferrooxidans SW2, an Fe(II) oxidoreductase involved in photoferrotrophy.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 68, 2012
|
|
4AWE
 
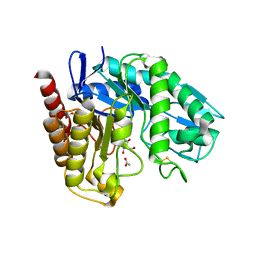 | | The Crystal Structure of Chrysonilia sitophila endo-beta-D-1,4- mannanase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goncalves, A.M.D, Silva, C.S, De Sanctis, D, Bento, I. | | Deposit date: | 2012-06-01 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Endo-Beta-D-1,4-Mannanase from Chrysonilia Sitophila Displays a Novel Loop Arrangement for Substrate Selectivity
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2IX1
 
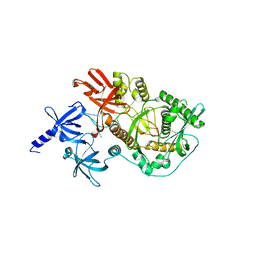 | | RNase II D209N mutant | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP *AP*AP*A)-3', EXORIBONUCLEASE 2, MAGNESIUM ION | | Authors: | Frazao, C, McVey, C.E, Amblar, M, Barbas, A, Vonrhein, C, Arraiano, C.M, Carrondo, M.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-10-05 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Unravelling the Dynamics of RNA Degradation by Ribonuclease II and its RNA-Bound Complex
Nature, 443, 2006
|
|
