1CLH
 
 | |
1TNS
 
 | |
2BB8
 
 | |
2EZH
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
2EZI
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, 30 STRUCTURES | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
1B69
 
 | |
1BB8
 
 | |
1TN9
 
 | |
1TNT
 
 | |
8DOV
 
 | | Crystal structure of the Shr Hemoglobin Interacting Domain 2 (HID2) in complex with Hemoglobin | | Descriptor: | GLYCEROL, Heme-binding protein Shr, Hemoglobin subunit alpha, ... | | Authors: | Macdonald, R, Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2022-07-14 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Shr receptor from Streptococcus pyogenes uses a cap and release mechanism to acquire heme-iron from human hemoglobin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6DKQ
 
 | | Crystal structure of the Shr Hemoglobin Interacting Domain 2 | | Descriptor: | Heme-binding protein Shr, SULFATE ION | | Authors: | Macdonald, R, Cascio, D, Collazo, M.J, Clubb, R.T. | | Deposit date: | 2018-05-30 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Streptococcus pyogenes Shr protein captures human hemoglobin using two structurally unique binding domains.
J.Biol.Chem., 293, 2018
|
|
7SJY
 
 | |
7K7F
 
 | |
8SQX
 
 | |
8SMU
 
 | | Integral fusion of the HtaA CR2 domain from Corynebacterium diphtheriae within EGFP | | Descriptor: | CHLORIDE ION, GLYCEROL, HtaACR2 integral fusion within enhanced green fluorescent protein, ... | | Authors: | Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2023-04-26 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development and atomic structure of a new fluorescence-based sensor to probe heme transfer in bacterial pathogens.
J.Inorg.Biochem., 249, 2023
|
|
7MPK
 
 | | Crystal structure of TagA with UDP-GlcNAc | | Descriptor: | N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Martinez, O.E, Cascio, D, Clubb, R.T. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Insight into the molecular basis of substrate recognition by the wall teichoic acid glycosyltransferase TagA.
J.Biol.Chem., 298, 2021
|
|
7N41
 
 | | Crystal structure of TagA with UDP-ManNAc | | Descriptor: | (2R,3S,4R,5S,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase | | Authors: | Martinez, O.E, Cascio, D, Clubb, R.T. | | Deposit date: | 2021-06-03 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insight into the molecular basis of substrate recognition by the wall teichoic acid glycosyltransferase TagA.
J.Biol.Chem., 298, 2021
|
|
4LFD
 
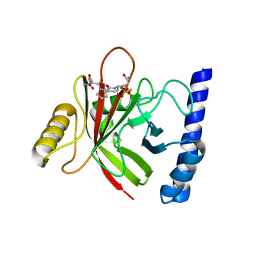 | | Staphylococcus aureus sortase B-substrate complex | | Descriptor: | (CBZ)NPQ(B27) PEPTIDE, SULFATE ION, Sortase B | | Authors: | Jacobitz, A.W, Sawaya, M.R, Yi, S.W, Amer, B.R, Huang, G.L, Nguyen, A.V, Jung, M.E, Clubb, R.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural and Computational Studies of the Staphylococcus aureus Sortase B-Substrate Complex Reveal a Substrate-stabilized Oxyanion Hole.
J.Biol.Chem., 289, 2014
|
|
4MYP
 
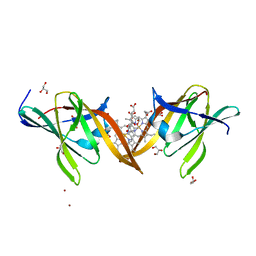 | | Structure of the central NEAT domain, N2, of the listerial Hbp2 protein complexed with heme | | Descriptor: | GLYCEROL, Iron-regulated surface determinant protein A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Malmirchegini, G.R, Sawaya, M.R, Clubb, R.T. | | Deposit date: | 2013-09-27 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Mechanism of Hemin Capture by Hbp2, the Hemoglobin-binding Hemophore from Listeria monocytogenes.
J.Biol.Chem., 289, 2014
|
|
4NLA
 
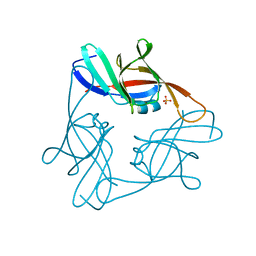 | | Structure of the central NEAT domain, N2, of the listerial Hbp2 protein, apo form | | Descriptor: | Iron-regulated surface determinant protein A, SULFATE ION | | Authors: | Malmirchegini, G.R, Sawaya, M.R, Clubb, R.T. | | Deposit date: | 2013-11-14 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel Mechanism of Hemin Capture by Hbp2, the Hemoglobin-binding Hemophore from Listeria monocytogenes.
J.Biol.Chem., 289, 2014
|
|
2WCC
 
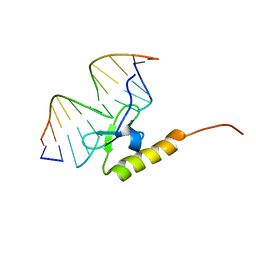 | | phage lambda IntDBD1-64 complex with p prime 2 DNA | | Descriptor: | DNA (5'-D(*DCP*DGP*DAP*DGP*DTP*DCP*DAP *DAP*DAP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DTP*DTP*DTP*DGP *DAP*DCP*DTP*DGP*DC)-3'), INTEGRASE | | Authors: | Fadeev, E.A, Sam, M.D, Clubb, R.T. | | Deposit date: | 2009-03-11 | | Release date: | 2009-04-07 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Amino-Terminal Domain of the Lambda Integrase Protein in Complex with DNA: Immobilization of a Flexible Tail Facilitates Beta- Sheet Recognition of the Major Groove.
J.Mol.Biol., 388, 2009
|
|
5CUW
 
 | |
5WB4
 
 | | Crystal structure of the TarA wall teichoic acid glycosyltransferase | | Descriptor: | N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase, SULFATE ION | | Authors: | Kattke, M.D, Cascio, D, Sawaya, M.R, Clubb, R.T. | | Deposit date: | 2017-06-27 | | Release date: | 2019-01-16 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of TagA, a novel membrane-associated glycosyltransferase that produces wall teichoic acids in pathogenic bacteria.
Plos Pathog., 15, 2019
|
|
5WFG
 
 | | Crystal structure of the TarA wall teichoic acid glycosyltransferase bound to UDP | | Descriptor: | N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Kattke, M.D, Cascio, D, Sawaya, M.R, Clubb, R.T. | | Deposit date: | 2017-07-11 | | Release date: | 2019-01-16 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of TagA, a novel membrane-associated glycosyltransferase that produces wall teichoic acids in pathogenic bacteria.
Plos Pathog., 15, 2019
|
|
5V8C
 
 | | LytR-Csp2A-Psr enzyme from Actinomyces oris | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Amer, B.R, Sawaya, M.R, Liauw, B, Fu, J.Y, Ton-That, H, Clubb, R.T. | | Deposit date: | 2017-03-21 | | Release date: | 2018-04-11 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure and Mechanism of LcpA, a Phosphotransferase That Mediates Glycosylation of a Gram-Positive Bacterial Cell Wall-Anchored Protein.
MBio, 10, 2019
|
|
