2LT2
 
 | |
2FCD
 
 | | Solution structure of N-lobe Myosin Light Chain from Saccharomices cerevisiae | | Descriptor: | Myosin light chain 1 | | Authors: | Cicero, D.O, Pennestri, M, Contessa, G.M, Paci, M, Ragnini-Wilson, A, Melino, S. | | Deposit date: | 2005-12-12 | | Release date: | 2006-11-07 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction of the myosin light chain Mlc1p with the myosin V Myo2p IQ motifs.
J.Biol.Chem., 282, 2007
|
|
2F1E
 
 | | Solution structure of ApaG protein | | Descriptor: | Protein apaG | | Authors: | Contessa, G, Pertinhez, T.A, Spisni, A, Paci, M, Farah, C.S, Cicero, D.O. | | Deposit date: | 2005-11-14 | | Release date: | 2006-10-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ApaG from Xanthomonas axonopodis pv. citri reveals a fibronectin-3 fold.
Proteins, 67, 2007
|
|
2FCE
 
 | | Solution structure of C-lobe Myosin Light Chain from Saccharomices cerevisiae | | Descriptor: | Myosin light chain 1 | | Authors: | Cicero, D.O, Pennestri, M, Contessa, G.M, Paci, M, Ragnini-Wilson, A, Melino, S. | | Deposit date: | 2005-12-12 | | Release date: | 2006-11-07 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction of the myosin light chain Mlc1p with the myosin V Myo2p IQ motifs.
J.Biol.Chem., 282, 2007
|
|
1BT7
 
 | | THE SOLUTION NMR STRUCTURE OF THE N-TERMINAL PROTEASE DOMAIN OF THE HEPATITIS C VIRUS (HCV) NS3-PROTEIN, FROM BK STRAIN, 20 STRUCTURES | | Descriptor: | NS3 SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Nardi, M.C, Steinkuhler, C, Cortese, R, De Francesco, R, Bazzo, R. | | Deposit date: | 1998-09-01 | | Release date: | 1999-06-22 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism.
J.Mol.Biol., 289, 1999
|
|
1SY9
 
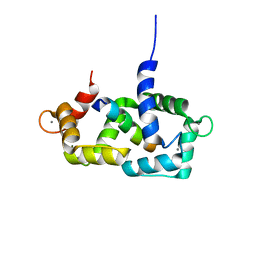 | | Structure of calmodulin complexed with a fragment of the olfactory CNG channel | | Descriptor: | CALCIUM ION, CALMODULIN, Cyclic-nucleotide-gated olfactory channel | | Authors: | Contessa, G.M, Orsale, M, Melino, S, Torre, V, Paci, M, Desideri, A, Cicero, D.O. | | Deposit date: | 2004-04-01 | | Release date: | 2005-04-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of calmodulin complexed with an olfactory CNG channel fragment and role of the central linker: residual dipolar couplings to evaluate calmodulin binding modes outside the kinase family.
J.Biomol.Nmr, 31, 2005
|
|
1DXW
 
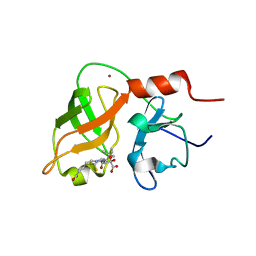 | | structure of hetero complex of non structural protein (NS) of hepatitis C virus (HCV) and synthetic peptidic compound | | Descriptor: | N-(tert-butoxycarbonyl)-L-alpha-glutamyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Cordier, F, Narjes, F, Gerlach, B, Sambucini, S, Grzesiek, S, Matassa, V.G, Defrancesco, R, Bazzo, R. | | Deposit date: | 2000-01-17 | | Release date: | 2001-01-12 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Inhibitor Binding Induces Active Site Stabilisation of the Hcv Ns3 Protein Serine Protease Domain
Embo J., 19, 2000
|
|
8B1L
 
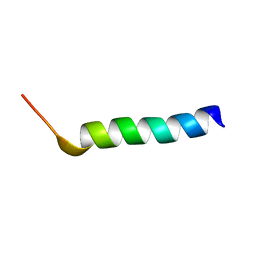 | |
6FE6
 
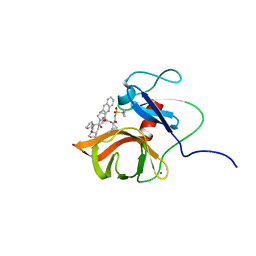 | | Solution structure of a last generation P2-P4 macrocyclic inhibitor | | Descriptor: | (3aR,7S,10S,12R,24aR)-7-cyclopentyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-5,8-dioxo-1,2,3,3a,5,6,7,8,11,12,20,21,22,23,24,24a-hexadecahydro-10H-9,12-methanocyclopenta[18,19][1,10,3,6]dioxadiazacyclononadecino[12,11-b]quinoline-10-carboxamide, Non-structural 3 protease, ZINC ION | | Authors: | Gallo, M, Eliseo, T, Cicero, D.O. | | Deposit date: | 2017-12-29 | | Release date: | 2019-01-30 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a last generation macrocyclic inhibitor. Hepatitis C virus NS3 protease complex: when S prime region occupancy is not enough to stabilize the protein conformation in the absence of NS4A.
To Be Published
|
|
2OII
 
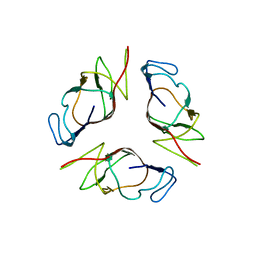 | | Structure of EMILIN-1 C1q-like domain | | Descriptor: | EMILIN-1 | | Authors: | Verdone, G, Colebrooke, S.A, Corazza, A, Cicero, D.O, Eliseo, T, Viglino, P, Campbell, I.D, Colombatti, A, Esposito, G. | | Deposit date: | 2007-01-11 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the C-terminal domain of EMILIN-1
To be Published
|
|
2K1Q
 
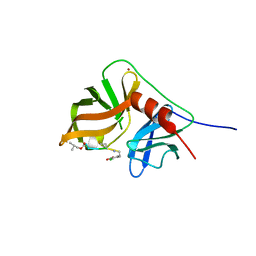 | | NMR structure of hepatitis c virus ns3 serine protease complexed with the non-covalently bound phenethylamide inhibitor | | Descriptor: | NS3 PROTEASE, PHENETHYLAMIDE, ZINC ION | | Authors: | Eliseo, T, Gallo, M, Pennestri, M, Bazzo, R, Cicero, D.O. | | Deposit date: | 2008-03-13 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Binding of a noncovalent inhibitor exploiting the S' region stabilizes the hepatitis C virus NS3 protease conformation in the absence of cofactor.
J.Mol.Biol., 385, 2009
|
|
2JN3
 
 | | NMR structure of cl-BABP complexed to chenodeoxycholic acid | | Descriptor: | CHENODEOXYCHOLIC ACID, Fatty acid-binding protein, liver | | Authors: | Eliseo, T, Ragona, L, Catalano, M, Assfalf, M, Paci, M, Zetta, L, Molinari, H, Cicero, D.O. | | Deposit date: | 2006-12-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic determinants of ligand binding in the ternary complex of chicken liver bile acid binding protein with two bile salts revealed by NMR
Biochemistry, 46, 2007
|
|
2K6L
 
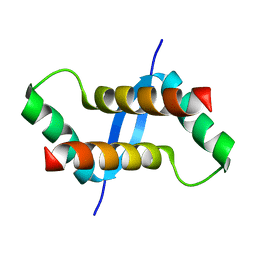 | | The solution structure of XACb0070 from Xanthonomas axonopodis pv citri reveals this new protein is a member of the RHH family of transcriptional repressors | | Descriptor: | Putative uncharacterized protein | | Authors: | Gallo, M, Cicero, D.O, Amata, I, Eliseo, T, Paci, M, Spisni, A, Ferrari, E, Pertinhez, T.A, Farah, C.S. | | Deposit date: | 2008-07-11 | | Release date: | 2009-06-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure reveals that XACb0070 from the plant pathogen Xanthomonas citri belongs to the RHH superfamily of bacterial DNA-binding proteins
To be Published
|
|
2KQA
 
 | | The solution structure of the fungal elicitor Cerato-Platanin | | Descriptor: | Cerato-platanin | | Authors: | Oliveira, A.L, Gallo, M, Pazzagli, L, Cappugi, G, Scala, A, Cicero, D.O, Pantera, B, Spisni, A, Benedetti, C.E, Pertinhez, T.A. | | Deposit date: | 2009-11-03 | | Release date: | 2011-03-23 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the fungal elicitor Cerato-Platanin
To be Published
|
|
1NXN
 
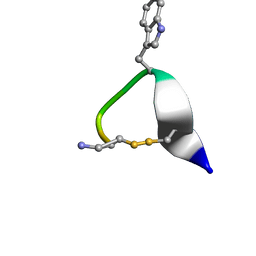 | | SOLUTION STRUCTURE OF CONTRYPHAN-VN | | Descriptor: | CONTRYPHAN-VN, MAJOR FORM (CIS CONFORMER) | | Authors: | Eliseo, T, Cicero, D.O, Polticelli, F, Schinina, M.E, Massilia, G.R, Paci, M, Ascenzi, P. | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-04 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cyclic peptide contryphan-Vn, a Ca2+-dependent K+ channel modulator
Biopolymers, 74, 2004
|
|
1R8P
 
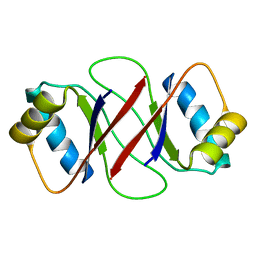 | | HPV-16 E2C solution structure | | Descriptor: | Regulatory protein E2 | | Authors: | Nadra, A.D, Eliseo, T, Cicero, D.O. | | Deposit date: | 2003-10-28 | | Release date: | 2004-11-23 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HPV-16 E2 DNA binding domain, a transcriptional regulator with a dimeric beta-barrel fold
J.Biomol.NMR, 30, 2004
|
|
1RG6
 
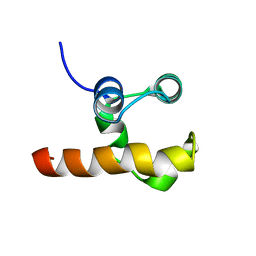 | | Solution structure of the C-terminal domain of p63 | | Descriptor: | second splice variant p63 | | Authors: | Cadot, B, Candi, E, Cicero, D.O, Desideri, A, Mele, S, Melino, G, Paci, M. | | Deposit date: | 2003-11-11 | | Release date: | 2004-11-23 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of p63
To be Published
|
|
4OA3
 
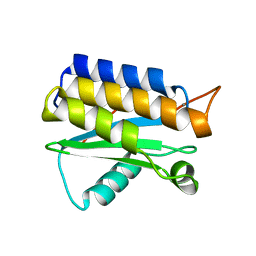 | | Crystal structure of the BA42 protein from BIZIONIA ARGENTINENSIS | | Descriptor: | CALCIUM ION, PROTEIN BA42 | | Authors: | Otero, L.H, Klinke, S, Aran, M, Pellizza, L, Goldbaum, F.A, Cicero, D. | | Deposit date: | 2014-01-03 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Solution and crystal structure of BA42, a protein from the Antarctic bacterium Bizionia argentinensis comprised of a stand-alone TPM domain.
Proteins, 82, 2014
|
|
3T50
 
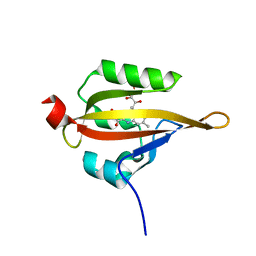 | |
2MPB
 
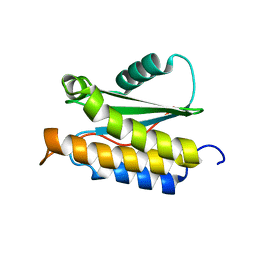 | | NMR structure of BA42 protein from the psychrophilic bacteria Bizionia argentinensis sp. nov | | Descriptor: | BA42 | | Authors: | Cicero, D, Aran, M, Smal, C, Pellizza, L, Gallo, M. | | Deposit date: | 2014-05-14 | | Release date: | 2014-08-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution and crystal structure of BA42, a protein from the Antarctic bacterium Bizionia argentinensis comprised of a stand-alone TPM domain.
Proteins, 82, 2014
|
|
2KA3
 
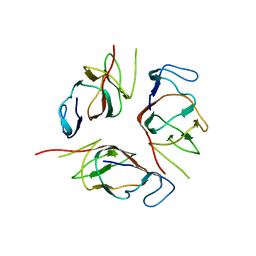 | | Structure of EMILIN-1 C1Q-like domain | | Descriptor: | EMILIN-1 | | Authors: | Verdone, G, Corazza, A, Colebrooke, S.A, Cicero, D.O, Eliseo, T, Boyd, J, Doliana, R, Fogolari, F, Viglino, P, Colombatti, A, Campbell, I.D, Esposito, G. | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR-based homology model for the solution structure of the C-terminal globular domain of EMILIN1
J.Biomol.Nmr, 43, 2009
|
|
1MZI
 
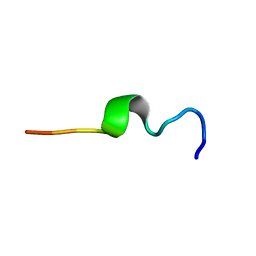 | | Solution ensemble structures of HIV-1 gp41 2F5 mAb epitope | | Descriptor: | 2F5 epitope of HIV-1 gp41 fusion protein | | Authors: | Barbato, G, Bianchi, E, Ingallinella, P, Hurni, W.H, Miller, M.D, Ciliberto, G, Cortese, R, Bazzo, R, Shiver, J.W, Pessi, A. | | Deposit date: | 2002-10-08 | | Release date: | 2003-09-02 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion.
J.Mol.Biol., 330, 2003
|
|
