1M3H
 
 | | Crystal Structure of Hogg1 D268E Mutant with Product Oligonucleotide | | Descriptor: | 5'-D(P*GP*CP*GP*TP*CP*CP*AP*(DDX))-3', 5'-D(P*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*TP*AP*CP*C)-3', ... | | Authors: | Chung, S.J, Verdine, G.L. | | Deposit date: | 2002-06-27 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of End Products Resulting from Lesion Processing by a DNA Glycosylase/Lyase
Chem.Biol., 11, 2004
|
|
1M3Q
 
 | |
1MQ0
 
 | | Crystal Structure of Human Cytidine Deaminase | | Descriptor: | 1-BETA-RIBOFURANOSYL-1,3-DIAZEPINONE, Cytidine Deaminase, ZINC ION | | Authors: | Chung, S.J, Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-09-13 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human cytidine deaminase bound to a potent inhibitor
J.Med.Chem., 48, 2005
|
|
4DCP
 
 | | Crystal Structure of caspase 3, L168F mutant | | Descriptor: | Caspase Inhibitor AC-DEVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Chung, S.J, Kang, H.J, Kim, S.J. | | Deposit date: | 2012-01-18 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insight into the role of the leucine residue on the L2 loop in the catalytic activity of caspases 3 and 7
Biosci.Rep., 32, 2012
|
|
4DCO
 
 | | Crystal Structure of caspase 3, L168Y mutant | | Descriptor: | Caspase Inhibitor AC-DEVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Chung, S.J, Kang, H.J, Kim, S.J. | | Deposit date: | 2012-01-18 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insight into the role of the leucine residue on the L2 loop in the catalytic activity of caspases 3 and 7
Biosci.Rep., 32, 2012
|
|
4DCJ
 
 | | Crystal structure of caspase 3, L168D mutant | | Descriptor: | Caspase Inhibitor AC-DEVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Chung, S.J, Kang, H.J, Kim, S.J. | | Deposit date: | 2012-01-17 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insight into the role of the leucine residue on the L2 loop in the catalytic activity of caspases 3 and 7
Biosci.Rep., 32, 2012
|
|
3B97
 
 | | Crystal Structure of human Enolase 1 | | Descriptor: | Alpha-enolase, MAGNESIUM ION, SULFATE ION | | Authors: | Kang, H.J, Jung, S.K, Kim, S.J, Chung, S.J. | | Deposit date: | 2007-11-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human alpha-enolase (hENO1), a multifunctional glycolytic enzyme.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4HQR
 
 | | Crystal Structure of mutant form of Caspase-7 | | Descriptor: | Ac-Asp-Glu-Val-Asp-Aldehyde, Caspase-7 | | Authors: | Lee, Y, Kang, H.J, Bae, K.-H, Kim, S.J, Chung, S.J. | | Deposit date: | 2012-10-26 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural asymmetry of procaspase-7 bound to a specific inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HQ0
 
 | | Crystal Structure of mutant form of Caspase-7 | | Descriptor: | Caspase-7 | | Authors: | Lee, Y, Kang, H.J, Bae, K.-H, Kim, S.J, Chung, S.J. | | Deposit date: | 2012-10-25 | | Release date: | 2013-09-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural asymmetry of procaspase-7 bound to a specific inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1HDU
 
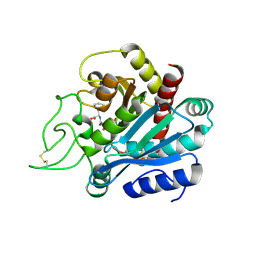 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with aminocarbonylphenylalanine at 1.75 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(AMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1HEE
 
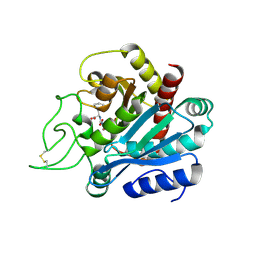 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with L-N-hydroxyaminocarbonyl phenylalanine at 2.3 A | | Descriptor: | CARBOXYPEPTIDASE A, L-[(N-HYDROXYAMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-22 | | Release date: | 2001-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1HDQ
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with D-N-hydroxyaminocarbonyl phenylalanine at 2.3 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(N-HYDROXYAMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1N39
 
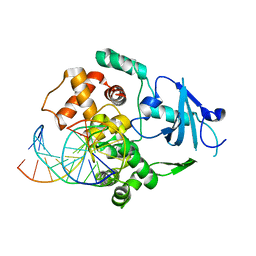 | | Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase | | Descriptor: | CALCIUM ION, DNA complement strand, DNA inhibitor strand, ... | | Authors: | Norman, D.P, Chung, S.J, Verdine, G.L. | | Deposit date: | 2002-10-25 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase
Biochemistry, 42, 2003
|
|
1N3A
 
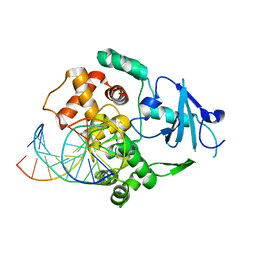 | | Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase | | Descriptor: | CALCIUM ION, DNA complement strand, DNA inhibitor strand, ... | | Authors: | Norman, D.P, Chung, S.J, Verdine, G.L. | | Deposit date: | 2002-10-25 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase
Biochemistry, 42, 2003
|
|
1N3C
 
 | | Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase | | Descriptor: | 8-oxoG-containing DNA, CALCIUM ION, DNA complement strand, ... | | Authors: | Norman, D.P, Chung, S.J, Verdine, G.L. | | Deposit date: | 2002-10-25 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase
Biochemistry, 42, 2003
|
|
4E89
 
 | | Crystal Structure of RnaseH from gammaretrovirus | | Descriptor: | CADMIUM ION, MAGNESIUM ION, RNase H | | Authors: | Kim, J.H, Kim, S.J. | | Deposit date: | 2012-03-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of xenotropic murine leukaemia virus-related virus (XMRV) ribonuclease H
Biosci.Rep., 32, 2012
|
|
6JKG
 
 | | The NAD+-free form of human NSDHL | | Descriptor: | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
6JKH
 
 | | The NAD+-bound form of human NSDHL | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
3HB1
 
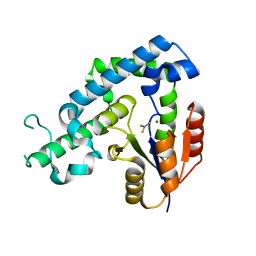 | | Crystal structure of ed-eya2 complexed with Alf3 | | Descriptor: | ALUMINUM FLUORIDE, Eyes absent homolog 2 (Drosophila), MAGNESIUM ION | | Authors: | Jung, S.K, Jeong, D.G, Ryu, S.E, Kim, S.J. | | Deposit date: | 2009-05-03 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of ED-Eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor
Faseb J., 24, 2010
|
|
3HB0
 
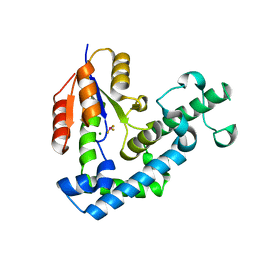 | | Structure of edeya2 complexed with bef3 | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Eyes absent homolog 2 (Drosophila), MAGNESIUM ION | | Authors: | Jung, S.K, Jeong, D.G, Ryu, S.E, Kim, S.J. | | Deposit date: | 2009-05-03 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of ED-Eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor
Faseb J., 24, 2010
|
|
4IKC
 
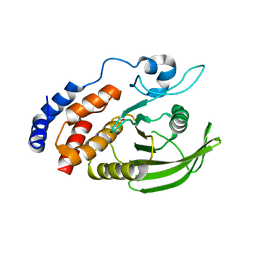 | | Crystal Structure of catalytic domain of PTPRQ | | Descriptor: | CHLORIDE ION, Phosphotidylinositol phosphatase PTPRQ, SULFATE ION | | Authors: | Yu, K.R, Ryu, S.E, Kim, S.J. | | Deposit date: | 2012-12-26 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis for the dephosphorylating activity of PTPRQ towards phosphatidylinositide substrates
Acta Crystallogr.,Sect.D, 69, 2013
|
|
