3QI0
 
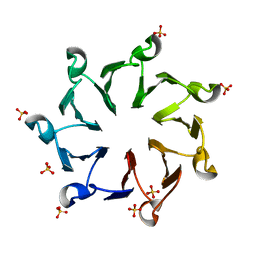 | | Structural, thermodynamic and kinetic analysis of the picomolar binding affinity interaction of the beta-lactamase inhibitor protein-II (BLIP-II) with class A beta-lactamases | | Descriptor: | Beta-lactamase inhibitory protein II, SULFATE ION | | Authors: | Brown, N.G, Chow, D.C, Sankaran, B, Zwart, P, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2011-01-26 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the binding forces driving the tight interactions between beta-lactamase inhibitory protein-II (BLIP-II) and class A beta-lactamases.
J.Biol.Chem., 286, 2011
|
|
3QHY
 
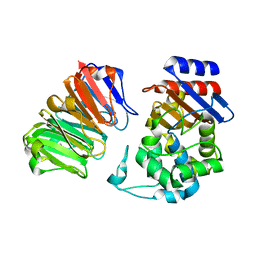 | | Structural, thermodynamic and kinetic analysis of the picomolar binding affinity interaction of the beta-lactamase inhibitor protein-II (BLIP-II) with class A beta-lactamases | | Descriptor: | Beta-lactamase, Beta-lactamase inhibitory protein II | | Authors: | Brown, N.G, Chow, D.C, Sankaran, B, Zwart, P, Prasad, B.V.V, Palzkill, T, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2011-01-26 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Analysis of the binding forces driving the tight interactions between beta-lactamase inhibitory protein-II (BLIP-II) and class A beta-lactamases.
J.Biol.Chem., 286, 2011
|
|
1P9M
 
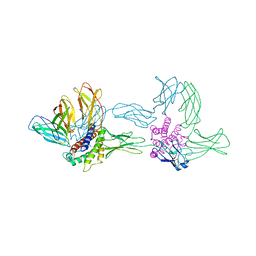 | | Crystal structure of the hexameric human IL-6/IL-6 alpha receptor/gp130 complex | | Descriptor: | Interleukin-6, Interleukin-6 receptor alpha chain, Interleukin-6 receptor beta chain | | Authors: | Boulanger, M.J, Chow, D.C, Brevnova, E.E, Garcia, K.C. | | Deposit date: | 2003-05-12 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Hexameric Structure and Assembly of the Interleukin-6/IL-6 alpha-Receptor/gp130 Complex
Science, 300, 2003
|
|
1R5V
 
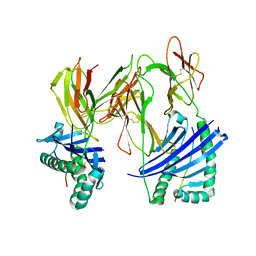 | | Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T-cell activation | | Descriptor: | H-2 class II histocompatibility antigen, E-K alpha chain, MHC H2-IE-beta, ... | | Authors: | Krogsgaard, M, Prado, N, Adams, E.J, He, X.L, Chow, D.C, Wilson, D.B, Garcia, K.C, Davis, M.M. | | Deposit date: | 2003-10-13 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation.
Mol.Cell, 12, 2003
|
|
1R5W
 
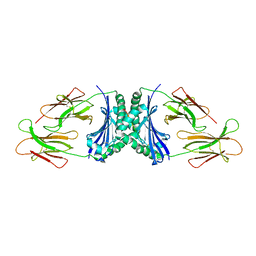 | | Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T-cell activation | | Descriptor: | H-2 class II histocompatibility antigen, E-K alpha chain, MHC H2-IE-beta, ... | | Authors: | Krogsgaard, M, Prado, N, Adams, E, He, X, Chow, D.C, Wilson, D.B, Garcia, K.C, Davis, M.M. | | Deposit date: | 2003-10-13 | | Release date: | 2004-03-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation
Mol.Cell, 12, 2003
|
|
4RVA
 
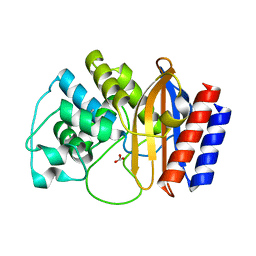 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for deacylation | | Descriptor: | BICARBONATE ION, Beta-lactamase TEM | | Authors: | Stojanoski, V, Chow, D.-C, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4397 Å) | | Cite: | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
4RX3
 
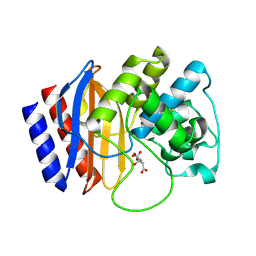 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for catalysis | | Descriptor: | Beta-lactamase TEM, CITRATE ANION | | Authors: | Stojanoski, V, Chow, D, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2014-12-08 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
4RX2
 
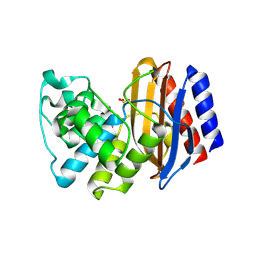 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for catalysis | | Descriptor: | Beta-lactamase TEM, SULFATE ION | | Authors: | Stojanoski, V, Chow, D, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2014-12-08 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.315 Å) | | Cite: | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
3C7V
 
 | |
3C7U
 
 | |
4S2M
 
 | | Crystal Structure of OXA-163 complexed with iodide in the active site | | Descriptor: | Beta-lactamase, IODIDE ION | | Authors: | Stojanoski, V, Hu, L, Palzkill, T.G, Prasad, B. | | Deposit date: | 2015-01-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural Basis for Different Substrate Profiles of Two Closely Related Class D beta-Lactamases and Their Inhibition by Halogens.
Biochemistry, 54, 2015
|
|
4S2L
 
 | | Crystal Structure of OXA-163 beta-lactamase | | Descriptor: | Beta-lactamase, SODIUM ION | | Authors: | Stojanoski, V, Liya, H, Palzkill, T.G, Prasad, B, Sankaran, B. | | Deposit date: | 2015-01-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for Different Substrate Profiles of Two Closely Related Class D beta-Lactamases and Their Inhibition by Halogens.
Biochemistry, 54, 2015
|
|
2LJL
 
 | |
1JDN
 
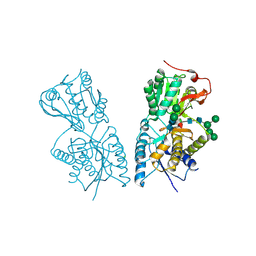 | | Crystal Structure of Hormone Receptor | | Descriptor: | ATRIAL NATRIURETIC PEPTIDE CLEARANCE RECEPTOR, CHLORIDE ION, beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, X.-L, Chow, D.-C, Martick, M.M, Garcia, K.C. | | Deposit date: | 2001-06-14 | | Release date: | 2001-09-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric activation of a spring-loaded natriuretic peptide receptor dimer by hormone.
Science, 293, 2001
|
|
1JDP
 
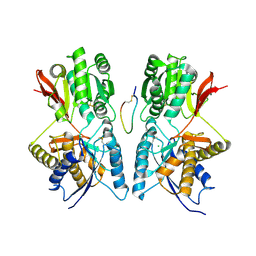 | | Crystal Structure of Hormone/Receptor Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATRIAL NATRIURETIC PEPTIDE CLEARANCE RECEPTOR, ... | | Authors: | He, X.-L, Chow, D.-C, Martick, M.M, Garcia, K.C. | | Deposit date: | 2001-06-14 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric activation of a spring-loaded natriuretic peptide receptor dimer by hormone.
Science, 293, 2001
|
|
3OCP
 
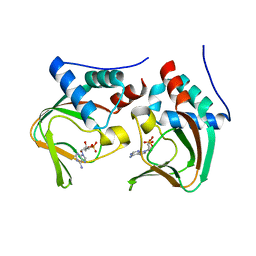 | | Crystal structure of cAMP bound cGMP-dependent protein kinase(92-227) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PRKG1 protein | | Authors: | Kim, J.J, Huang, G, Kwon, T.K, Zwart, P, Headd, J, Kim, C. | | Deposit date: | 2010-08-10 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Co-Crystal Structures of PKG Ibeta (92-227) with cGMP and cAMP Reveal the Molecular Details of Cyclic-Nucleotide Binding
Plos One, 6, 2011
|
|
3OD0
 
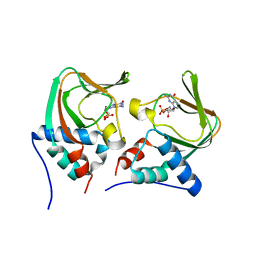 | | Crystal structure of cGMP bound cGMP-dependent protein kinase(92-227) | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, PRKG1 protein | | Authors: | Kim, J.J, Huang, G, Kwon, T.K, Zwart, P, Headd, J, Kim, C. | | Deposit date: | 2010-08-10 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Co-Crystal Structures of PKG Ibeta (92-227) with cGMP and cAMP Reveal the Molecular Details of Cyclic-Nucleotide Binding
Plos One, 6, 2011
|
|
3OGJ
 
 | | Crystal structure of partial apo (92-227) of cGMP-dependent protein kinase | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PHOSPHATE ION, PRKG1 protein | | Authors: | Kim, J.J, Huang, G, Kwon, T.K, Zwart, P, Headd, J, Kim, C. | | Deposit date: | 2010-08-16 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Co-Crystal Structures of PKG Ibeta (92-227) with cGMP and cAMP Reveal the Molecular Details of Cyclic-Nucleotide Binding
Plos One, 6, 2011
|
|
4G0A
 
 | |
4G0J
 
 | |
