2FA5
 
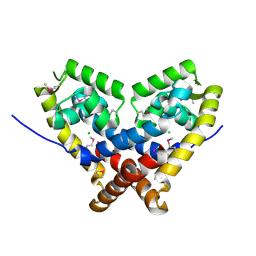 | | The crystal structure of an unliganded multiple antibiotic-resistance repressor (MarR) from Xanthomonas campestris | | Descriptor: | CHLORIDE ION, transcriptional regulator marR/emrR family | | Authors: | Chin, K.H, Tu, Z.L, Li, J.N, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2005-12-06 | | Release date: | 2006-11-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of XC1739: a putative multiple antibiotic-resistance repressor (MarR) from Xanthomonas campestris at 1.8 A resolution
Proteins, 65, 2006
|
|
2FUJ
 
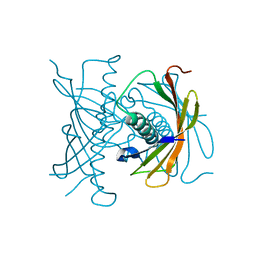 | |
2FUK
 
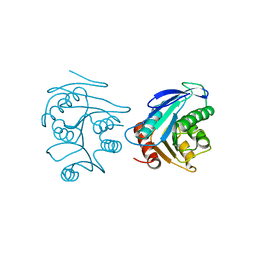 | | Crystal structure of XC6422 from Xanthomonas campestris: a member of a/b serine hydrolase without lid at 1.6 resolution | | Descriptor: | XC6422 protein | | Authors: | Yang, C.Y, Chin, K.H, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2006-01-27 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of XC6422 from Xanthomonas campestris at 1.6 A resolution: a small serine alpha/beta-hydrolase
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1C11
 
 | | INTERCALATED D(TCCCGTTTCCA) DIMER, NMR, 7 STRUCTURES | | Descriptor: | DNA (5'-D(*TP*CP*CP*CP*GP*TP*TP*TP*CP*CP*A)-3') | | Authors: | Gallego, J, Chou, S.H, Reid, B.R. | | Deposit date: | 1998-07-15 | | Release date: | 1998-07-22 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Centromeric pyrimidine strands fold into an intercalated motif by forming a double hairpin with a novel T:G:G:T tetrad: solution structure of the d(TCCCGTTTCCA) dimer.
J.Mol.Biol., 273, 1997
|
|
4YP1
 
 | | Misting with CDA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Stimulator of interferon genes protein | | Authors: | Chin, K.H, Chen, C.K, Tu, Z.I, Chou, S.H. | | Deposit date: | 2015-03-12 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Insights into the Distinct Binding Mode of Cyclic Di-AMP with SaCpaA_RCK.
Biochemistry, 54, 2015
|
|
4YS2
 
 | | RCK domain with CDA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Na+/H+ antiporter-like protein | | Authors: | Chin, K.H, Chou, S.H. | | Deposit date: | 2015-03-16 | | Release date: | 2016-04-27 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | RCK domain with CDA
To Be Published
|
|
2ZE3
 
 | |
2GBZ
 
 | | The Crystal Structure of XC847 from Xanthomonas campestris: a 3-5 Oligoribonuclease of DnaQ fold family with a Novel Opposingly-Shifted Helix | | Descriptor: | MAGNESIUM ION, Oligoribonuclease | | Authors: | Chin, K.H, Yang, C.Y, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2006-03-12 | | Release date: | 2007-01-16 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of XC847 from Xanthomonas campestris: a 3'-5' oligoribonuclease of DnaQ fold family with a novel opposingly shifted helix
Proteins, 65, 2006
|
|
1L3M
 
 | | The Solution Structure of [d(CGC)r(amamam)d(TTTGCG)]2 | | Descriptor: | 5'-D(*CP*GP*C)-R(P*(A39)P*(A39)P*(A39))-D(P*TP*TP*TP*GP*CP*G)-3' | | Authors: | Tsao, Y.P, Wang, L.Y, Hsu, S.T, Jain, M.L, Chou, S.H, Huang, W.C, Cheng, J.W. | | Deposit date: | 2002-02-28 | | Release date: | 2002-04-03 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of [d(CGC)r(amamam)d(TTTGCG)]2.
J.Biomol.NMR, 21, 2001
|
|
1P0U
 
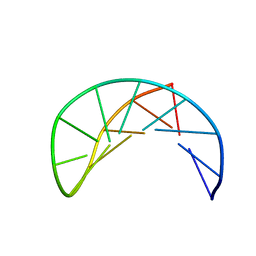 | | Sheared G/C Base Pair | | Descriptor: | 5'-D(*GP*CP*AP*TP*CP*GP*AP*CP*GP*AP*TP*GP*C)-3' | | Authors: | Chin, K.H, Chou, S.H. | | Deposit date: | 2003-04-11 | | Release date: | 2003-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Sheared-type G(anti).C(syn) Base-pair: A Unique d(GXC) Loop Closure Motif.
J.Mol.Biol., 329, 2003
|
|
4KBY
 
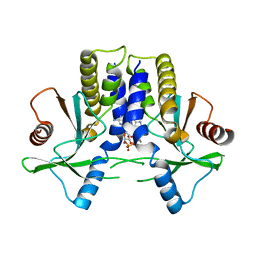 | | mSTING/c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Stimulator of interferon genes protein | | Authors: | Chin, K.H, Su, Y.C, Tu, J.L, Chou, S.H. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4KC0
 
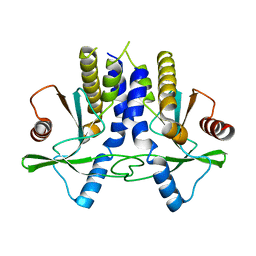 | | mSTING | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Chin, K.H, Su, Y.C, Tu, J.L, Chou, S.H. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3DSG
 
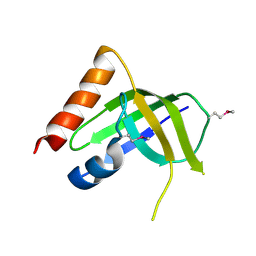 | | XC1028 from Xanthomonas campestris Adopts a PilZ Domain-like Structure Yet with Trivial c-di-GMP Binding Activity | | Descriptor: | Type IV fimbriae assembly protein | | Authors: | Li, T.N, Chin, K.H, Liu, J.H, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2008-07-12 | | Release date: | 2009-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | XC1028 from Xanthomonas campestris adopts a PilZ domain-like structure without a c-di-GMP switch.
Proteins, 75, 2009
|
|
3IWZ
 
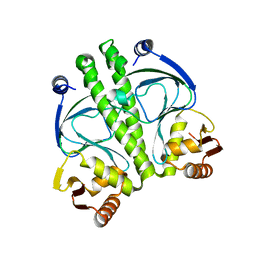 | | The c-di-GMP Responsive Global Regulator CLP Links Cell-Cell Signaling to Virulence Gene Expression in Xanthomonas campestris | | Descriptor: | Catabolite activation-like protein | | Authors: | Chin, K.H, Tu, Z.L, Tseng, Y.H, Dow, J.M, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2009-09-03 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris.
J.Mol.Biol., 396, 2010
|
|
2GSC
 
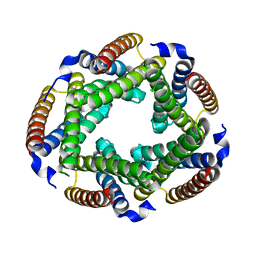 | | Crystal Structure of the Conserved Hypothetical Cytosolic Protein Xcc0516 from Xanthomonas campestris | | Descriptor: | Putative uncharacterized protein XCC0516 | | Authors: | Lin, L.Y, Ching, C.L, Chin, K.H, Chou, S.H, Chan, N.L. | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the conserved hypothetical cytosolic protein Xcc0516 from Xanthomonas campestris reveals a novel quaternary structure assembled by five four-helix bundles.
Proteins, 65, 2006
|
|
1BJH
 
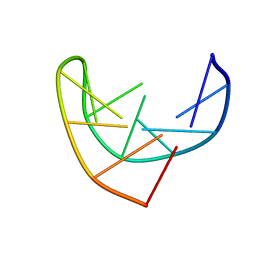 | | HAIRPIN LOOPS CONSISTING OF SINGLE ADENINE RESIDUES CLOSED BY SHEARED A(DOT)A AND G(DOT)G PAIRS FORMED BY THE DNA TRIPLETS AAA AND GAG: SOLUTION STRUCTURE OF THE D(GTACAAAGTAC) HAIRPIN, NMR, 16 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*AP*AP*AP*GP*TP*AP*C)-3') | | Authors: | Chou, S.-H, Zhu, L, Gao, Z, Cheng, J.-W, Reid, B.R. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Hairpin loops consisting of single adenine residues closed by sheared A.A and G.G pairs formed by the DNA triplets AAA and GAG: solution structure of the d(GTACAAAGTAC) hairpin.
J.Mol.Biol., 264, 1996
|
|
1D69
 
 | |
175D
 
 | |
103D
 
 | |
1L1V
 
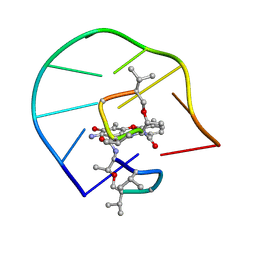 | | UNUSUAL ACTD/DNA_TA COMPLEX STRUCTURE | | Descriptor: | 5'-D(*GP*TP*CP*AP*CP*CP*GP*AP*C)-3', ACTINOMYCIN D | | Authors: | Chou, S.-H, Chin, K.-H, Chen, F.-M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-03-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Looped Out and Perpendicular: Deformation of Watson-Crick Base Pair Associated with Actinomycin D Binding.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1D68
 
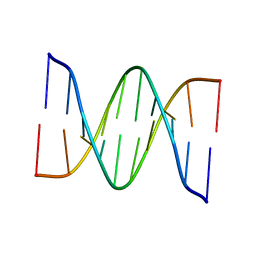 | |
1CHV
 
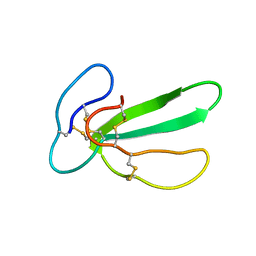 | |
4S05
 
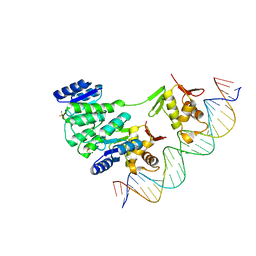 | | Crystal structure of Klebsiella pneumoniae PmrA in complex with PmrA box DNA | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA (26-MER), DNA-binding transcriptional regulator BasR, ... | | Authors: | Hsiao, C.D, Weng, T.H, Li, Y.C. | | Deposit date: | 2014-12-30 | | Release date: | 2015-11-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA.
Nat Commun, 6, 2015
|
|
4S04
 
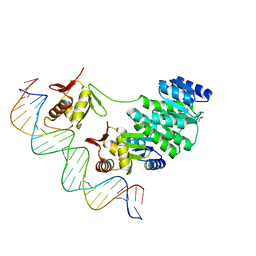 | | Crystal structure of Klebsiella pneumoniae PmrA in complex with PmrA box DNA | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA (25-MER), DNA-binding transcriptional regulator BasR, ... | | Authors: | Hsiao, C.D, Weng, T.H, Li, Y.C. | | Deposit date: | 2014-12-30 | | Release date: | 2015-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA.
Nat Commun, 6, 2015
|
|
7X6R
 
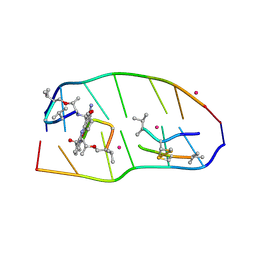 | | Crystal structure of actinomycin D-echinomycin-d(AGCACGT/ACGGGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, Actinomycin D, COBALT (II) ION, ... | | Authors: | Kao, S.H, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-08 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
