8I4R
 
 | |
8I4P
 
 | |
7VRX
 
 | | Pad-1 in the absence of substrate | | Descriptor: | Aminotransferase, SULFATE ION | | Authors: | Choi, M, Rhee, S. | | Deposit date: | 2021-10-25 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96634674 Å) | | Cite: | Structural and biochemical basis for the substrate specificity of Pad-1, an indole-3-pyruvic acid aminotransferase in auxin homeostasis.
J.Struct.Biol., 214, 2022
|
|
1YP0
 
 | | Structure of the steroidogenic factor-1 ligand binding domain bound to phospholipid and a SHP peptide motif | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor subfamily 0, group B, ... | | Authors: | Li, Y, Choi, M, Cavey, G, Daugherty, J, Suino, K, Kovach, A, Bingham, N, Kliewer, S, Xu, H. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1.
Mol.Cell, 17, 2005
|
|
1ZGY
 
 | | Structural and Biochemical Basis for Selective Repression of the Orphan Nuclear Receptor LRH-1 by SHP | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), Nuclear receptor subfamily 0, ... | | Authors: | Li, Y, Choi, M, Suino, K, Kovach, A, Daugherty, J, Kliewer, S.A, Xu, H.E. | | Deposit date: | 2005-04-22 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZH7
 
 | | Structural and Biochemical Basis for Selective Repression of the Orphan Nuclear Receptor LRH-1 by SHP | | Descriptor: | Orphan nuclear receptor NR5A2, nuclear receptor subfamily 0, group B, ... | | Authors: | Li, Y, Choi, M, Suino, K, Kovach, A, Daugherty, J, Kliewer, S.A, Xu, H.E. | | Deposit date: | 2005-04-22 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3IE9
 
 | | Structure of oxidized M98L mutant of amicyanin | | Descriptor: | ACETATE ION, Amicyanin, CHLORIDE ION, ... | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining the role of the axial ligand of the type 1 copper site in amicyanin by replacement of methionine with leucine.
Biochemistry, 48, 2009
|
|
3IEA
 
 | | Structure of reduced M98L mutant of amicyanin | | Descriptor: | ACETATE ION, Amicyanin, CHLORIDE ION, ... | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Defining the role of the axial ligand of the type 1 copper site in amicyanin by replacement of methionine with leucine.
Biochemistry, 48, 2009
|
|
3PLY
 
 | | Structure of Oxidized P96G Mutant of Amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2010-11-15 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Proline 96 of the copper ligand loop of amicyanin regulates electron transfer from methylamine dehydrogenase by positioning other residues at the protein-protein interface.
Biochemistry, 50, 2011
|
|
6XFP
 
 | | Crystal Structure of BRAF kinase domain bound to Belvarafenib | | Descriptor: | 4-amino-N-{1-[(3-chloro-2-fluorophenyl)amino]-6-methylisoquinolin-5-yl}thieno[3,2-d]pyrimidine-7-carboxamide, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Yin, J, Sudhamsu, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma.
Nature, 594, 2021
|
|
4QVE
 
 | | Crystal structure of Bcl-xL in complex with BID BH3 domain | | Descriptor: | Bcl-2-like protein 1, IMIDAZOLE, Peptide from BH3-interacting domain death agonist | | Authors: | Sreekanth, R, Yoon, H.S. | | Deposit date: | 2014-07-14 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Bh3 induced conformational changes in Bcl-Xl revealed by crystal structure and comparative analysis.
Proteins, 83, 2015
|
|
8HG9
 
 | |
4Y5R
 
 | | Crystal Structure of a T67A MauG/pre-Methylamine Dehydrogenase Complex | | Descriptor: | CALCIUM ION, HEME C, Methylamine dehydrogenase heavy chain, ... | | Authors: | Li, C, Wilmot, C.M. | | Deposit date: | 2015-02-11 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A T67A mutation in the proximal pocket of the high-spin heme of MauG stabilizes formation of a mixed-valent Fe(II)/Fe(III) state and enhances charge resonance stabilization of the bis-Fe(IV) state.
Biochim.Biophys.Acta, 1847, 2015
|
|
3TH5
 
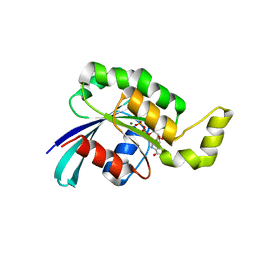 | | Crystal structure of wild-type RAC1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
4DHF
 
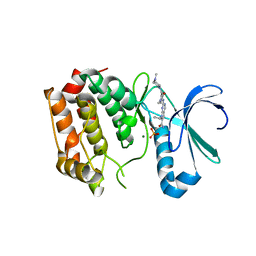 | | Structure of Aurora A mutant bound to Biogenidec cpd 15 | | Descriptor: | 7-cyclopentyl-2-({1-methyl-5-[(4-methylpiperazin-1-yl)carbonyl]-1H-pyrrol-3-yl}amino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | Silvian, L, Marcotte, D.J. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of 2,6,7-trisubstituted-7H-pyrrolo[2,3-d]pyrimidines as Aurora kinases inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5H4V
 
 | |
3SBE
 
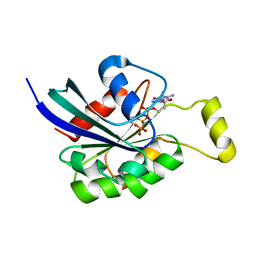 | | Crystal structure of RAC1 P29S mutant | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-06-03 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
3SBD
 
 | | Crystal structure of RAC1 P29S mutant | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-06-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
3KCF
 
 | |
4P5S
 
 | | Structure of reduced W45Y mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (I) ION | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The sole tryptophan of amicyanin enhances its thermal stability but does not influence the electronic properties of the type 1 copper site.
Arch.Biochem.Biophys., 550-551, 2014
|
|
4P5R
 
 | | Structure of oxidized W45Y mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION, SODIUM ION | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The sole tryptophan of amicyanin enhances its thermal stability but does not influence the electronic properties of the type 1 copper site.
Arch.Biochem.Biophys., 550-551, 2014
|
|
4PPI
 
 | | Crystal structure of Bcl-xL hexamer | | Descriptor: | Bcl-2-like protein 1, GLYCEROL | | Authors: | Sreekanth, R, Yoon, H.S. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Structural transition in Bcl-xL and its potential association with mitochondrial calcium ion transport
Sci Rep, 5, 2015
|
|
4QVF
 
 | |
3RYM
 
 | | Structure of Oxidized M98K mutant of Amicyanin | | Descriptor: | Amicyanin, ZINC ION | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2011-05-11 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7039 Å) | | Cite: | Replacement of the axial copper ligand methionine with lysine in amicyanin converts it to a zinc-binding protein that no longer binds copper.
J.Inorg.Biochem., 105, 2011
|
|
