8YJX
 
 | |
3X2Y
 
 | | Crystal structure of metallo-beta-lactamase H8A from Thermotoga maritima | | Descriptor: | NICKEL (II) ION, UPF0173 metal-dependent hydrolase TM_1162 | | Authors: | Choi, H.J, Kim, H.J, Matsuura, A, Mikami, B, Yoon, H.J, Lee, H.H. | | Deposit date: | 2015-01-07 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of metallo-beta-lactamase H8A from Thermotoga maritima
To be Published
|
|
3X30
 
 | | Crystal structure of metallo-beta-lactamase from Thermotoga maritima | | Descriptor: | MANGANESE (II) ION, NICKEL (II) ION, UPF0173 metal-dependent hydrolase TM_1162 | | Authors: | Choi, H.J, Kim, H.J, Matsuura, A, Mikami, B, Yoon, H.J, Lee, H.H. | | Deposit date: | 2015-01-07 | | Release date: | 2016-02-17 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Crystal structure of metallo-beta-lactamase from Thermotoga maritima
To be Published
|
|
3X2X
 
 | | Crystal structure of metallo-beta-lactamase H48A from Thermotoga maritima | | Descriptor: | MANGANESE (II) ION, UPF0173 metal-dependent hydrolase TM_1162 | | Authors: | Choi, H.J, Kim, H.J, Matsuura, A, Mikami, B, Yoon, H.J, Lee, H.H. | | Deposit date: | 2015-01-07 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Crystal structure of metallo-beta-lactamase H48A from Thermotoga maritima
To be Published
|
|
3X2Z
 
 | | Crystal structure of metallo-beta-lactamase in complex with nickel from Thermotoga maritima | | Descriptor: | NICKEL (II) ION, UPF0173 metal-dependent hydrolase TM_1162 | | Authors: | Choi, H.J, Kim, H.J, Matsuura, A, Mikami, B, Yoon, H.J, Lee, H.H. | | Deposit date: | 2015-01-07 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of metallo-beta-lactamase in complex with nickel from Thermotoga maritima
To be Published
|
|
1LM7
 
 | | Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure | | Descriptor: | subdomain of Desmoplakin Carboxy-Terminal domain (DPCT) | | Authors: | Choi, H.J, Park-Snyder, S, Pascoe, L.T, Green, K.J, Weis, W.I. | | Deposit date: | 2002-04-30 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure.
Nat.Struct.Biol., 9, 2002
|
|
1LM5
 
 | | Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure | | Descriptor: | subdomain of Desmoplakin Carboxy-Terminal domain (DPCT) | | Authors: | Choi, H.J, Park-Snyder, S, Pascoe, L.T, Green, K.J, Weis, W.I. | | Deposit date: | 2002-04-30 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure.
Nat.Struct.Biol., 9, 2002
|
|
1XM9
 
 | |
4X98
 
 | | Immunoglobulin Fc heterodimer variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Seok, S.H, Choi, H.J, Kim, Y.J, Seo, M.D, Kim, Y.S. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Crystal structures of immunoglobulin Fc heterodimers reveal the molecular basis for heterodimer formation.
Mol.Immunol., 65, 2015
|
|
4X99
 
 | | Immunoglobulin Fc heterodimers variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Seok, S.H, Choi, H.J, Kim, Y.J, Seo, M.D, Kim, Y.S. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structures of immunoglobulin Fc heterodimers reveal the molecular basis for heterodimer formation.
Mol.Immunol., 65, 2015
|
|
7C0J
 
 | | Crystal structure of chimeric mutant of GH5 in complex with Z-DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), Histone H5,Double-stranded RNA-specific adenosine deaminase | | Authors: | Choi, H.J, Park, C.H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dual conformational recognition by Z-DNA binding protein is important for the B-Z transition process.
Nucleic Acids Res., 48, 2020
|
|
7C0I
 
 | | Crystal structure of chimeric mutant of E3L in complex with Z-DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), Double-stranded RNA-binding protein,Double-stranded RNA-specific adenosine deaminase, SULFATE ION | | Authors: | Choi, H.J, Park, C.H, Kim, J.S. | | Deposit date: | 2020-05-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dual conformational recognition by Z-DNA binding protein is important for the B-Z transition process.
Nucleic Acids Res., 48, 2020
|
|
2RH1
 
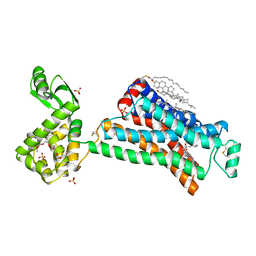 | | High resolution crystal structure of human B2-adrenergic G protein-coupled receptor. | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Cherezov, V, Rosenbaum, D.M, Hanson, M.A, Rasmussen, S.G.F, Thian, F.S, Kobilka, T.S, Choi, H.J, Kuhn, P, Weis, W.I, Kobilka, B.K, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2007-10-05 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor.
Science, 318, 2007
|
|
2R4S
 
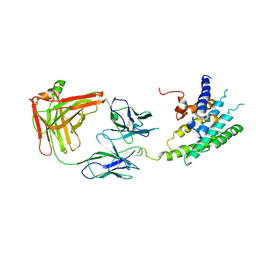 | | Crystal structure of the human beta2 adrenoceptor | | Descriptor: | Beta-2 adrenergic receptor, antibody for beta2 adrenoceptor, heavy chain, ... | | Authors: | Rasmussen, S.G.F, Choi, H.J, Rosenbaum, D.M, Kobilka, T.S, Thian, F.S, Edwards, P.C, Burghammer, M, Ratnala, V.R, Sanishvili, R, Fischetti, R.F, Schertler, G.F, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the human beta2 adrenergic G-protein-coupled receptor.
Nature, 450, 2007
|
|
2R4R
 
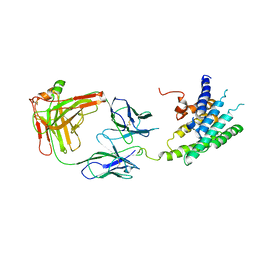 | | Crystal structure of the human beta2 adrenoceptor | | Descriptor: | Beta-2 adrenergic receptor, antibody for beta2 adrenoceptor, heavy chain, ... | | Authors: | Rasmussen, S.G.F, Choi, H.J, Rosenbaum, D.M, Kobilka, T.S, Thian, F.S, Edwards, P.C, Burghammer, M, Ratnala, V.R, Sanishvili, R, Fischetti, R.F, Schertler, G.F, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the human beta2 adrenergic G-protein-coupled receptor.
Nature, 450, 2007
|
|
5H5M
 
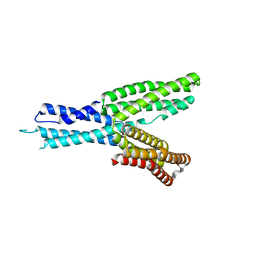 | | Crystal structure of HMP-1 M domain | | Descriptor: | Alpha-catenin-like protein hmp-1 | | Authors: | Kang, H, Bang, I, Weis, W.I, Choi, H.J. | | Deposit date: | 2016-11-08 | | Release date: | 2017-03-29 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of Caenorhabditis elegans alpha-catenin reveals constitutive binding to beta-catenin and F-actin
J. Biol. Chem., 292, 2017
|
|
5XA5
 
 | | Crystal structure of HMP-1-HMP-2 complex | | Descriptor: | Alpha-catenin-like protein hmp-1, Beta-catenin-like protein hmp-2 | | Authors: | Shao, X, Kang, H, Weis, W.I, Hardin, J, Choi, H.J. | | Deposit date: | 2017-03-11 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cell-cell adhesion in metazoans relies on evolutionarily conserved features of the alpha-catenin· beta-catenin-binding interface.
J.Biol.Chem., 292, 2017
|
|
6KL7
 
 | | Beta-arrestin 1 mutant S13D/T275D | | Descriptor: | 1,2-ETHANEDIOL, BARIUM ION, Beta-arrestin-1 | | Authors: | Kang, H, Choi, H.J. | | Deposit date: | 2019-07-29 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Conformational Dynamics and Functional Implications of Phosphorylated beta-Arrestins.
Structure, 28, 2020
|
|
6L5H
 
 | |
6L5J
 
 | |
4R10
 
 | | A conserved phosphorylation switch controls the interaction between cadherin and beta-catenin in vitro and in vivo | | Descriptor: | 1,2-ETHANEDIOL, Cadherin-related hmr-1, Protein humpback-2, ... | | Authors: | Choi, H.-J, Loveless, T, Lynch, A, Bang, I, Hardin, J, Weis, W.I. | | Deposit date: | 2014-08-03 | | Release date: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Conserved Phosphorylation Switch Controls the Interaction between Cadherin and beta-Catenin In Vitro and In Vivo
Dev.Cell, 33, 2015
|
|
3IFQ
 
 | | Interction of plakoglobin and beta-catenin with desmosomal cadherins | | Descriptor: | E-cadherin, SULFATE ION, plakoglobin | | Authors: | Choi, H.-J, Gross, J.C, Pokutta, S, Weis, W.I. | | Deposit date: | 2009-07-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interactions of plakoglobin and beta-catenin with desmosomal cadherins: basis of selective exclusion of alpha- and beta-catenin from desmosomes.
J.Biol.Chem., 284, 2009
|
|
1PRX
 
 | | HORF6 A NOVEL HUMAN PEROXIDASE ENZYME | | Descriptor: | HORF6 | | Authors: | Choi, H.-J, Kang, S.W, Yang, C.-H, Rhee, S.G, Ryu, S.-E. | | Deposit date: | 1998-04-03 | | Release date: | 1998-06-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a novel human peroxidase enzyme at 2.0 A resolution.
Nat.Struct.Biol., 5, 1998
|
|
4R11
 
 | | A conserved phosphorylation switch controls the interaction between cadherin and beta-catenin in vitro and in vivo | | Descriptor: | Cadherin-related hmr-1, IODIDE ION, Protein humpback-2 | | Authors: | Choi, H.-J, Loveless, T, Lynch, A, Bang, I, Hardin, J, Weis, W.I. | | Deposit date: | 2014-08-03 | | Release date: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | A Conserved Phosphorylation Switch Controls the Interaction between Cadherin and beta-Catenin In Vitro and In Vivo
Dev.Cell, 33, 2015
|
|
4R0Z
 
 | | A conserved phosphorylation switch controls the interaction between cadherin and beta-catenin in vitro and in vivo | | Descriptor: | FORMIC ACID, Protein humpback-2 | | Authors: | Choi, H.-J, Loveless, T, Lynch, A, Bang, I, Hardin, J, Weis, W.I. | | Deposit date: | 2014-08-03 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | A Conserved Phosphorylation Switch Controls the Interaction between Cadherin and beta-Catenin In Vitro and In Vivo
Dev.Cell, 33, 2015
|
|
