4UBD
 
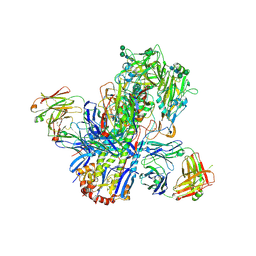 | | Crystal structure of a neutralizing human monoclonal antibody with 1968 H3 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Shore, D.A, Yang, H, Cho, M, Donis, R.O, Stevens, J. | | Deposit date: | 2014-08-12 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus.
Nat Commun, 6, 2015
|
|
7Y1H
 
 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | Descriptor: | 1-(5-chloranyl-4-methyl-benzimidazol-1-yl)-3-[(2R,3S)-3-oxidanylpiperidin-2-yl]propan-2-one, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | Deposit date: | 2022-06-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
7Y1W
 
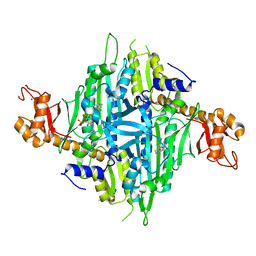 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | Descriptor: | (2R,3S)-2-[3-[4,5-bis(chloranyl)benzimidazol-1-yl]propyl]piperidin-3-ol, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
7Y28
 
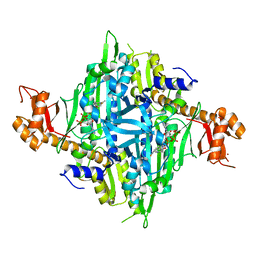 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | Descriptor: | 1-[6-(3-fluorophenyl)benzimidazol-1-yl]-3-[(2R,3S)-3-oxidanylpiperidin-2-yl]propan-2-one, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
7Y3S
 
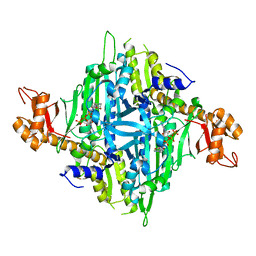 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | Descriptor: | 1-(6-bromanyl-7-methyl-imidazo[4,5-b]pyridin-3-yl)-3-[(2R,3S)-3-oxidanylpiperidin-2-yl]propan-2-one, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | Deposit date: | 2022-06-12 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
4R8W
 
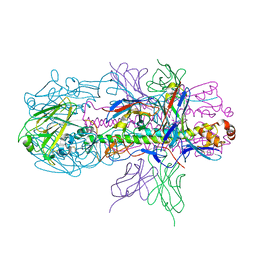 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a neutralizing antibody CT149 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of neutralizing antibody CT149, Hemagglutinin, ... | | Authors: | Wu, Y, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus
Nat Commun, 6, 2015
|
|
5JI1
 
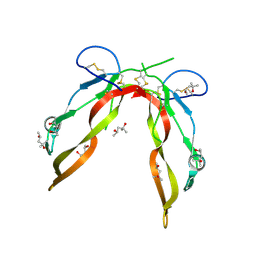 | | Crystal Structure of GDF8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Growth/differentiation factor 8 | | Authors: | Walker, R.G, Thompson, T.B. | | Deposit date: | 2016-04-21 | | Release date: | 2017-03-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for potency differences between GDF8 and GDF11.
BMC Biol., 15, 2017
|
|
2LYR
 
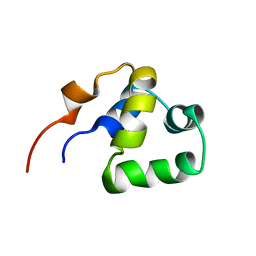 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYK
 
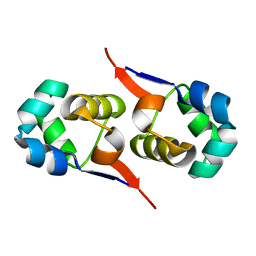 | | NOE-based 3D structure of the CylR2 homodimer at 270K (-3 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYL
 
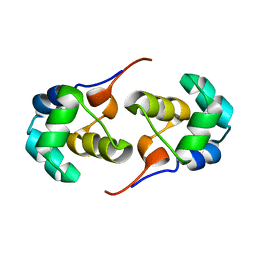 | | NOE-based 3D structure of the predissociated homodimer of CylR2 in equilibrium with monomer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYS
 
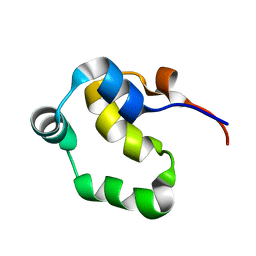 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 257K (-16 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYJ
 
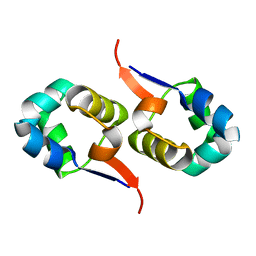 | | NOE-based 3D structure of the CylR2 homodimer at 298K | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Giller, K, Becker, S, Zweckstetter, M, Schwieters, C.D. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYP
 
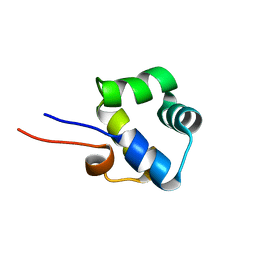 | | NOE-based 3D structure of the monomer of CylR2 in equilibrium with predissociated homodimer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYQ
 
 | | NOE-based 3D structure of the monomeric intermediate of CylR2 at 262K (-11 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
5JHW
 
 | |
5UHM
 
 | |
3K9A
 
 | | Crystal Structure of HIV gp41 with MPER | | Descriptor: | HIV glycoprotein gp41 | | Authors: | Shi, W, Han, D, Habte, H, Cho, M, Chance, M.R. | | Deposit date: | 2009-10-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of HIV gp41 with the membrane-proximal external region
J.Biol.Chem., 285, 2010
|
|
