2MF2
 
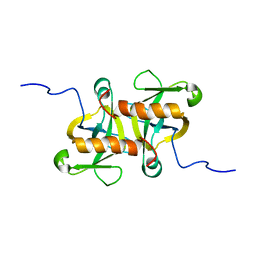 | |
4MZM
 
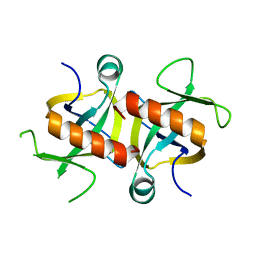 | | MazF from S. aureus crystal form I, P212121, 2.1 A | | Descriptor: | mRNA interferase MazF | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
4MZP
 
 | | MazF from S. aureus crystal form III, C2221, 2.7 A | | Descriptor: | MazF mRNA interferase | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
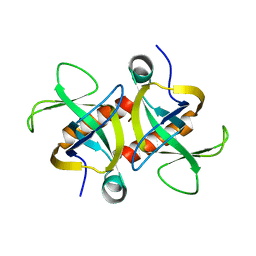
4MZT
 
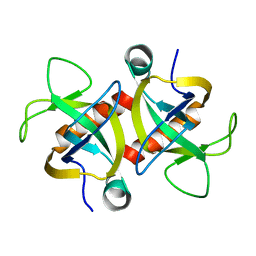 | | MazF from S. aureus crystal form II, C2221, 2.3 A | | Descriptor: | MazF mRNA interferase | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
6HV9
 
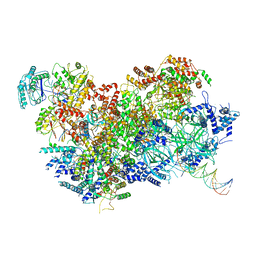 | | S. cerevisiae CMG-Pol epsilon-DNA | | Descriptor: | Cell division control protein 45, DNA (5'-D(*GP*CP*AP*GP*CP*CP*AP*CP*GP*CP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*GP*CP*GP*TP*GP*GP*CP*TP*GP*C)-3'), ... | | Authors: | Abid Ali, F, Purkiss, A.G, Cheung, A, Costa, A. | | Deposit date: | 2018-10-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (4.98 Å) | | Cite: | Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome.
Nat Commun, 9, 2018
|
|
6HV8
 
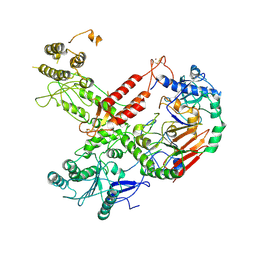 | | Cryo-EM structure of S. cerevisiae Polymerase epsilon deltacat mutant | | Descriptor: | DNA polymerase epsilon catalytic subunit A, DNA polymerase epsilon subunit B, ZINC ION | | Authors: | Goswami, P, Purkiss, A, Cheung, A, Costa, A. | | Deposit date: | 2018-10-10 | | Release date: | 2018-12-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome.
Nat Commun, 9, 2018
|
|
6T9K
 
 | | SAGA Core module | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6T9J
 
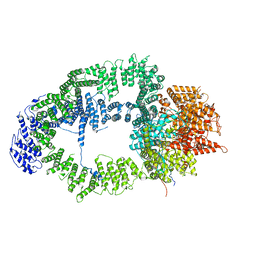 | | SAGA Tra1 module | | Descriptor: | Transcription factor SPT20, Transcription initiation factor TFIID subunit 12, Transcription-associated protein 1 | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6T9I
 
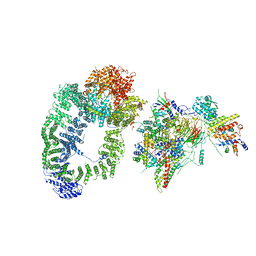 | | cryo-EM structure of transcription coactivator SAGA | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
3BRV
 
 | | NEMO/IKKb association domain structure | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit beta, NF-kappa-B essential modulator | | Authors: | Silvian, L.F. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a NEMO/IKK-Associating Domain Reveals Architecture of the Interaction Site.
Structure, 16, 2008
|
|
3BRT
 
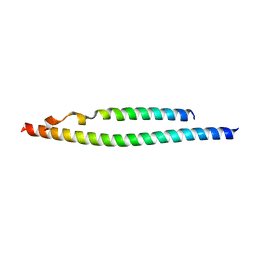 | | NEMO/IKK association domain structure | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit beta,Inhibitor of nuclear factor kappa-B kinase subunit alpha, NF-kappa-B essential modulator | | Authors: | Silvian, L.F. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a NEMO/IKK-Associating Domain Reveals Architecture of the Interaction Site.
Structure, 16, 2008
|
|
4BOC
 
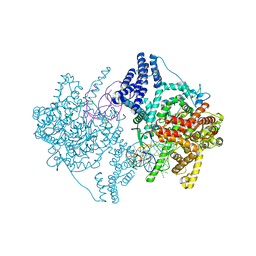 | | Structure of mitochondrial RNA polymerase elongation complex | | Descriptor: | 5'-D(*CP*AP*TP*GP*GP*GP*GP*TP*AP*AP*TP*TP*AP*TP *TP*TP*CP*GP*AP*CP*GP*CP*CP*AP*GP*AP*CP*G)-3', 5'-D(*CP*GP*TP*CP*TP*GP*GP*CP*GP*TP*GP*CP*GP*CP *GP*CP*CP*GP*CP*TP*AP*CP*CP*CP*CP*AP*TP*G)-3', 5'-R(*AP*GP*UP*CP*UP*GP*CP*GP*GP*CP*GP*CP*GP*CP)-3', ... | | Authors: | Schwinghammer, K, Cheung, A, Morozov, Y, Agaronyan, K, Temiakov, D, Cramer, P. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of Human Mitochondrial RNA Polymerase Elongation Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
7L9Y
 
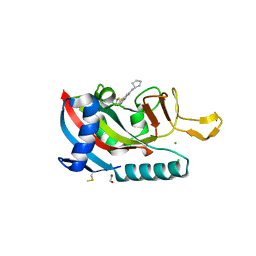 | | Human PARP14 (ARTD8), catalytic fragment in complex with RBN012042 | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopentylamino)-5-fluoro-2-{[(piperidin-4-yl)sulfanyl]methyl}quinazolin-4(3H)-one, CHLORIDE ION, ... | | Authors: | Dorsey, B.W, Swinger, K.K, Schenkel, L.B, Church, W.D, Perl, N.R, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2021-01-05 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Targeted Degradation of PARP14 Using a Heterobifunctional Small Molecule.
Chembiochem, 22, 2021
|
|
7LUN
 
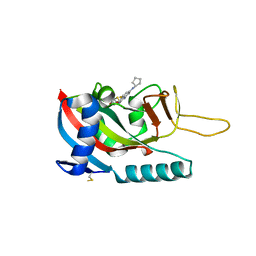 | | Human PARP14 (ARTD8), catalytic fragment in complex with RBN011980 | | Descriptor: | 7-(cyclopentylamino)-5-fluoro-2-{[(trans-4-hydroxycyclohexyl)sulfanyl]methyl}quinazolin-4(3H)-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dorsey, B.W, Swinger, K.K, Schenkel, L.B, Church, W.D, Perl, N.R, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Targeted Degradation of PARP14 Using a Heterobifunctional Small Molecule.
Chembiochem, 22, 2021
|
|
3UH4
 
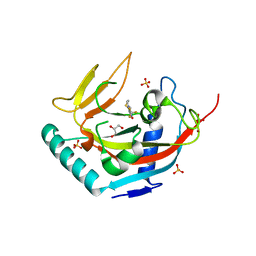 | | TANKYRASE-1 complexed with NVP-XAV939 | | Descriptor: | 2-[4-(trifluoromethyl)phenyl]-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-4-ol, GLYCEROL, SULFATE ION, ... | | Authors: | Stams, T, Kirby, C. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human tankyrase 1 in complex with small-molecule inhibitors PJ34 and XAV939.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3UH2
 
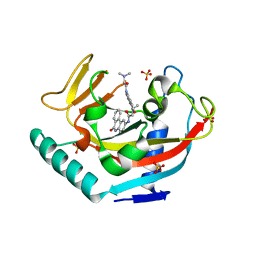 | | Tankyrase-1 in complexed with PJ34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, SULFATE ION, Tankyrase-1, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human tankyrase 1 in complex with small-molecule inhibitors PJ34 and XAV939.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3UDD
 
 | | Tankyrase-1 in complex with small molecule inhibitor | | Descriptor: | 3-(4-methoxyphenyl)-5-({[4-(4-methoxyphenyl)-5-methyl-4H-1,2,4-triazol-3-yl]sulfanyl}methyl)-1,2,4-oxadiazole, GLYCEROL, SULFATE ION, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | [1,2,4]triazol-3-ylsulfanylmethyl)-3-phenyl-[1,2,4]oxadiazoles: antagonists of the Wnt pathway that inhibit tankyrases 1 and 2 via novel adenosine pocket binding.
J.Med.Chem., 55, 2012
|
|
