2GAG
 
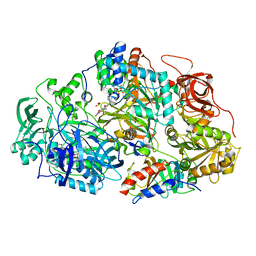 | | Heteroteterameric sarcosine: structure of a diflavin metaloenzyme at 1.85 a resolution | | Descriptor: | 2-FUROIC ACID, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chen, Z.W, Hassan-Abdulah, A, Zhao, G, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Heterotetrameric sarcosine oxidase: structure of a diflavin metalloenzyme at 1.85 a resolution.
J.Mol.Biol., 360, 2006
|
|
2GAH
 
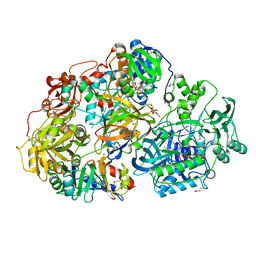 | | Heterotetrameric sarcosine: structure of a diflavin metaloenzyme at 1.85 a resolution | | Descriptor: | 2-FUROIC ACID, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chen, Z.W, Hassan-Abdulah, A, Zhao, G, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Heterotetrameric sarcosine oxidase: structure of a diflavin metalloenzyme at 1.85 a resolution.
J.Mol.Biol., 360, 2006
|
|
1FCD
 
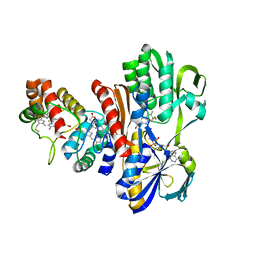 | | THE STRUCTURE OF FLAVOCYTOCHROME C SULFIDE DEHYDROGENASE FROM A PURPLE PHOTOTROPHIC BACTERIUM CHROMATIUM VINOSUM AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C SULFIDE DEHYDROGENASE (CYTOCHROME SUBUNIT), FLAVOCYTOCHROME C SULFIDE DEHYDROGENASE (FLAVIN-BINDING SUBUNIT), ... | | Authors: | Chen, Z.W, Koh, M, Van Driessche, G, Van Beeumen, J.J, Bartsch, R.G, Meyer, T.E, Cusanovich, M.A, Mathews, F.S. | | Deposit date: | 1994-08-18 | | Release date: | 1994-11-01 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium.
Science, 266, 1994
|
|
1YIQ
 
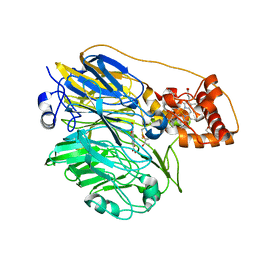 | | Molecular cloning and structural analysis of quinohemoprotein alcohol dehydrogenase ADHIIG from Pseudomonas putida HK5. Compariison to the other quinohemoprotein alcohol dehydrogenase ADHIIB found in the same microorganism. | | Descriptor: | CALCIUM ION, HEME C, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Toyama, H, Chen, Z.W, Fukumoto, M, Adachi, O, Matsushita, K, Mathews, F.S. | | Deposit date: | 2005-01-12 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular cloning and structural analysis of quinohemoprotein alcohol dehydrogenase ADH-IIG from Pseudomonas putida HK5
J.Mol.Biol., 352, 2005
|
|
1DII
 
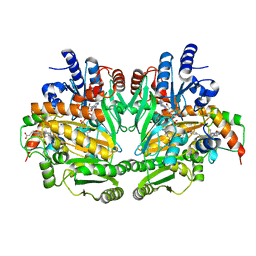 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE AT 2.5 A RESOLUTION | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | Authors: | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.N, McIntire, W.S. | | Deposit date: | 1999-11-29 | | Release date: | 1999-12-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
1DIQ
 
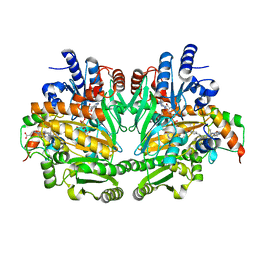 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE WITH SUBSTRATE BOUND | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | Authors: | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.S, McIntire, W.S. | | Deposit date: | 1999-11-29 | | Release date: | 1999-12-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
1MG2
 
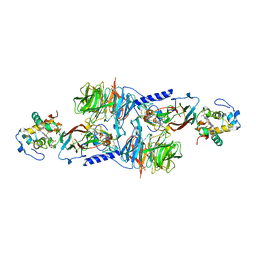 | | MUTATION OF ALPHA PHE55 OF METHYLAMINE DEHYDROGENASE ALTERS THE REORGANIZATION ENERGY AND ELECTRONIC COUPLING FOR ITS ELECTRON TRANSFER REACTION WITH AMICYANIN | | Descriptor: | Amicyanin, COPPER (II) ION, CYTOCHROME C-L, ... | | Authors: | Sun, D, Chen, Z.W, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2002-08-14 | | Release date: | 2002-12-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | MUTATION OF AlPHA PHE55 OF METHYLAMINE DEHYDROGENASE ALTERS THE REORGANIZATION ENERGY AND ELECTRONIC COUPLING FOR ITS ELECTRON TRANSFER REACTION WITH AMICYANIN
Biochemistry, 41, 2002
|
|
1MG3
 
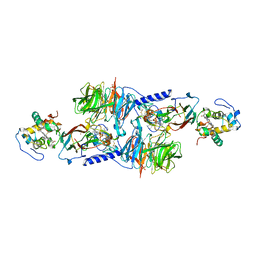 | | MUTATION OF ALPHA PHE55 OF METHYLAMINE DEHYDROGENASE ALTERS THE REORGANIZATION ENERGY AND ELECTRONIC COUPLING FOR ITS ELECTRON TRANSFER REACTION WITH AMICYANIN | | Descriptor: | Amicyanin, COPPER (II) ION, CYTOCHROME C-L, ... | | Authors: | Sun, D, Chen, Z.W, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2002-08-14 | | Release date: | 2002-12-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | MUTATION OF ALPHA PHE55 OF METHYLAMINE DEHYDROGENASE ALTERS THE REORGANIZATION ENERGY AND ELECTRONIC COUPLING FOR ITS ELECTRON TRANSFER REACTION WITH AMICYANIN
Biochemistry, 41, 2002
|
|
1SFQ
 
 | | Fast form of thrombin mutant R(77a)A bound to PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, ... | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-20 | | Release date: | 2004-06-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
1TB3
 
 | | Crystal Structure Analysis of Recombinant Rat Kidney Long-chain Hydroxy Acid Oxidase | | Descriptor: | ACETIC ACID, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 3 | | Authors: | Cunane, L.M, Barton, J.D, Chen, Z.W, Le, K.H.D, Amar, D, Lederer, F, Mathews, F.S. | | Deposit date: | 2004-05-19 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of Recombinant Rat Kidney Long Chain Hydroxy Acid Oxidase.
Biochemistry, 44, 2005
|
|
1SG8
 
 | | Crystal structure of the procoagulant fast form of thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, thrombin | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
1SGI
 
 | | Crystal structure of the anticoagulant slow form of thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, thrombin | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
1SHH
 
 | | Slow form of Thrombin Bound with PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, thrombin | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-25 | | Release date: | 2004-06-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
2HWL
 
 | | Crystal structure of thrombin in complex with fibrinogen gamma' peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen gamma' peptide, Prothrombin, ... | | Authors: | Pineda, A.O, Chen, Z.W, Marino, F, Mathews, F.S, Mosesson, M.W, Di Cera, E. | | Deposit date: | 2006-08-01 | | Release date: | 2006-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of thrombin in complex with fibrinogen gamma' peptide.
Biophys.Chem., 125, 2007
|
|
1KBJ
 
 | | Crystallographic Study of the Recombinant Flavin-binding Domain of Baker's Yeast Flavocytochrome b2: comparison with the Intact Wild-type Enzyme | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME B2, FLAVIN MONONUCLEOTIDE | | Authors: | Cunane, L.M, Barton, J.D, Chen, Z.W, Welsh, F.E, Chapman, S.K, Reid, G.A, Mathews, F.S. | | Deposit date: | 2001-11-06 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic study of the recombinant flavin-binding domain of Baker's yeast flavocytochrome b(2): comparison with the intact wild-type enzyme.
Biochemistry, 41, 2002
|
|
3SGZ
 
 | | High resolution crystal structure of rat long chain hydroxy acid oxidase in complex with the inhibitor 4-carboxy-5-[(4-chiorophenyl)sulfanyl]-1, 2, 3-thiadiazole. | | Descriptor: | 5-[(4-methylphenyl)sulfanyl]-1,2,3-thiadiazole-4-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 2 | | Authors: | Chen, Z, Vignaud, C, Jaafar, A, Gueritte, F, Guenard, D, Lederer, F, Mathews, F.S. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution crystal structure of rat long chain hydroxy acid oxidase in complex with the inhibitor 4-carboxy-5-[(4-chlorophenyl)sulfanyl]-1, 2, 3-thiadiazole. Implications for inhibitor specificity and drug design.
Biochimie, 94, 2012
|
|
1NWP
 
 | |
1NWO
 
 | |
1KV9
 
 | | Structure at 1.9 A Resolution of a Quinohemoprotein Alcohol Dehydrogenase from Pseudomonas putida HK5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETONE, CALCIUM ION, ... | | Authors: | Chen, Z.-W, Matsushita, K, Yamashita, T, Fujii, T, Toyama, H, Adachi, O, Bellamy, H.D, Mathews, F.S. | | Deposit date: | 2002-01-25 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure at 1.9 A resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas putida HK5.
Structure, 10, 2002
|
|
3NBJ
 
 | | Crystal Structure of Y305F mutant of the copper amine oxidase from Hansenula polymorpha expressed in yeast | | Descriptor: | COPPER (II) ION, PHOSPHATE ION, Peroxisomal primary amine oxidase | | Authors: | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2010-06-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
3N9H
 
 | | Crystal Structural of mutant Y305A in the copper amine oxidase from hansenula polymorpha | | Descriptor: | COPPER (II) ION, Peroxisomal primary amine oxidase | | Authors: | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2010-05-30 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
3NBB
 
 | | Crystal structure of mutant Y305F expressed in E. coli in the copper amine oxidase from hansenula polymorpha | | Descriptor: | COPPER (II) ION, Peroxisomal primary amine oxidase | | Authors: | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2010-06-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
1AAC
 
 | | AMICYANIN OXIDIZED, 1.31 ANGSTROMS | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Cunane, L.M, Chen, Z.-W, Durley, R.C.E, Mathews, F.S. | | Deposit date: | 1995-09-07 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | X-ray structure of the cupredoxin amicyanin, from Paracoccus denitrificans, refined at 1.31 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1BHG
 
 | | HUMAN BETA-GLUCURONIDASE AT 2.6 A RESOLUTION | | Descriptor: | BETA-GLUCURONIDASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Jain, S, Drendel, W.B. | | Deposit date: | 1996-03-04 | | Release date: | 1997-09-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure of human beta-glucuronidase reveals candidate lysosomal targeting and active-site motifs.
Nat.Struct.Biol., 3, 1996
|
|
1BXA
 
 | | AMICYANIN REDUCED, PH 4.4, 1.3 ANGSTROMS | | Descriptor: | COPPER (I) ION, PROTEIN (AMICYANIN) | | Authors: | Cunane, L.M, Chen, Z.W, Durley, R.C.E, Mathews, F.S. | | Deposit date: | 1998-10-01 | | Release date: | 1998-10-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for interprotein complex-dependent effects on the redox properties of amicyanin.
Biochemistry, 37, 1998
|
|
