1VYU
 
 | | Beta3 subunit of Voltage-gated Ca2+-channel | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Liang, T, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
1VYV
 
 | | beta4 subunit of Ca2+ channel | | Descriptor: | CALCIUM CHANNEL BETA-4SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Liang, T, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
1VYT
 
 | | beta3 subunit complexed with aid | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT, VOLTAGE-DEPENDENT L-TYPE CALCIUM CHANNEL ALPHA-1C SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Tong, L, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
3M78
 
 | |
3M7E
 
 | |
3M72
 
 | |
3M77
 
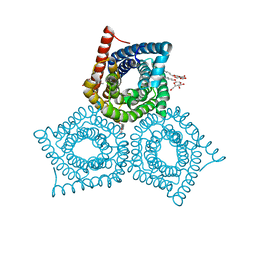 | |
3M7C
 
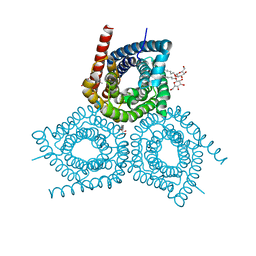 | |
3M70
 
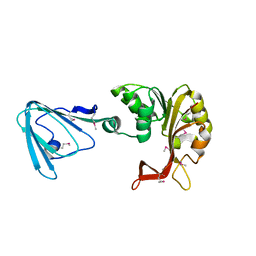 | |
3M7B
 
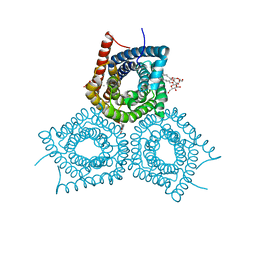 | |
2WDW
 
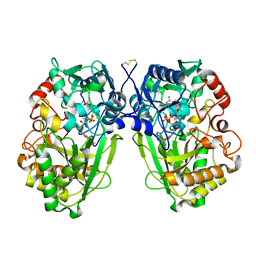 | | The Native Crystal Structure of the Primary Hexose Oxidase (Dbv29) in Antibiotic A40926 Biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PUTATIVE HEXOSE OXIDASE | | Authors: | Liu, Y.-C, Li, Y.-S, Lyu, S.-Y, Chen, Y.-H, Chan, H.-C, Huang, C.-J, Huang, Y.-T, Chen, G.-H, Chou, C.-C, Tsai, M.-D, Li, T.-L. | | Deposit date: | 2009-03-27 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Interception of Teicoplanin Oxidation Intermediates Yields New Antimicrobial Scaffolds
Nat.Chem.Biol., 7, 2011
|
|
4CDZ
 
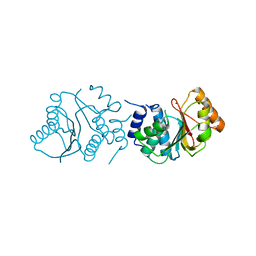 | | Crystal Structure of Spinosyn Rhamnosyl 4'-O-Methyltransferase SpnH from Saccharopolyspora spinosa | | Descriptor: | MAGNESIUM ION, O-METHYLTRANSFERASE | | Authors: | Lin, Y.-C, Huang, S.-P, Huang, B.-L, Chen, Y.-H, Chen, Y.-J, Chiu, H.-T. | | Deposit date: | 2013-11-08 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal Structure and Functional Insights of Spinosyn Rhamnosyl 4'-O-Methyltransferase Spnh from Saccharopolyspora Spinosa
To be Published
|
|
4CE0
 
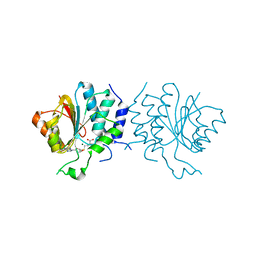 | | Crystal Structure of SAH-bound Spinosyn Rhamnosyl 4'-O- Methyltransferase SpnH from Saccharopolyspora spinosa | | Descriptor: | MAGNESIUM ION, O-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lin, Y.-C, Huang, S.-P, Huang, B.-L, Chen, Y.-H, Chen, Y.-J, Chiu, H.-T. | | Deposit date: | 2013-11-08 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure and Functional Insights of Spinosyn Rhamnosyl 4'-O-Methyltransferase Spnh from Saccharopolyspora Spinosa
To be Published
|
|
5I10
 
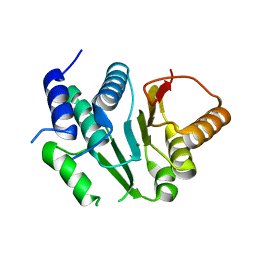 | | Crystal structure of spinosyn rhamnosyl 4'-O-methyltransferase spnh mutant T242Q from Saccharopolyspora Spinosa | | Descriptor: | MAGNESIUM ION, Probable O-methyltransferase | | Authors: | Lin, Y.-C, Huang, S.-P, Huang, B.-L, Chen, Y.-H, Chen, Y.-J, Chiu, H.-T. | | Deposit date: | 2016-02-05 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of spinosyn rhamnosyl 4'-O-methyltransferase spnh mutant T242Q from Saccharopolyspora Spinosa
To Be Published
|
|
3M74
 
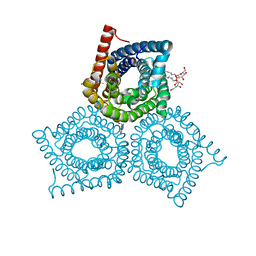 | |
3M7L
 
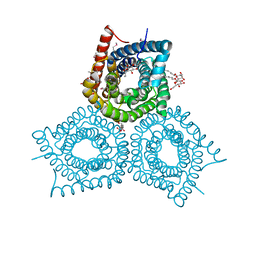 | |
3M71
 
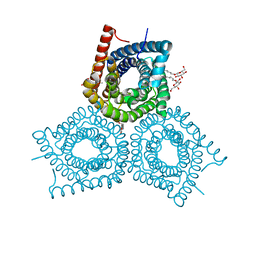 | | Crystal Structure of Plant SLAC1 homolog TehA | | Descriptor: | Tellurite resistance protein tehA homolog, octyl beta-D-glucopyranoside | | Authors: | Chen, Y.-H, Hu, L, Punta, M, Bruni, R, Hillerich, B, Kloss, B, Rost, B, Love, J, Siegelbaum, S.A, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2010-03-16 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Homologue structure of the SLAC1 anion channel for closing stomata in leaves.
Nature, 467, 2010
|
|
3M75
 
 | |
3M76
 
 | |
3M73
 
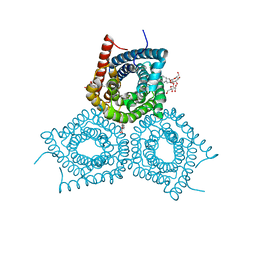 | |
3DEJ
 
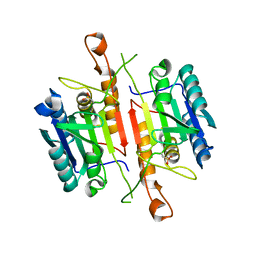 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | Descriptor: | (1S)-1-(3-chlorophenyl)-2-oxo-2-[(1,3,4-trioxo-1,2,3,4-tetrahydroisoquinolin-5-yl)amino]ethyl acetate, Caspase-3 | | Authors: | Wu, J, Du, J, Li, J, Ding, J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
3DEH
 
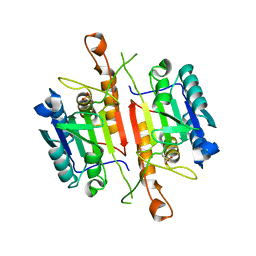 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | Descriptor: | Caspase-3, isoquinoline-1,3,4(2H)-trione | | Authors: | Wu, J, Du, J, Li, J, Ding, J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
3DEI
 
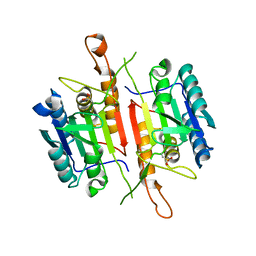 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | Descriptor: | (1S)-2-oxo-1-phenyl-2-[(1,3,4-trioxo-1,2,3,4-tetrahydroisoquinolin-5-yl)amino]ethyl acetate, Caspase-3 | | Authors: | Wu, J, Du, J, Li, J, Ding, J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
3DEK
 
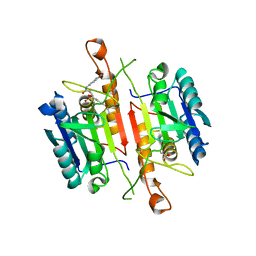 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | Descriptor: | Caspase-3, N-[3-(2-fluoroethoxy)phenyl]-N'-(1,3,4-trioxo-1,2,3,4-tetrahydroisoquinolin-6-yl)butanediamide | | Authors: | Wu, J, Du, J, Li, J, Ding, J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
