4ZHT
 
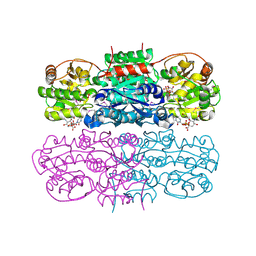 | | Crystal structure of UDP-GlcNAc 2-epimerase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-mannopyranose, Bifunctional UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase, CYTIDINE-5'-MONOPHOSPHATE-5-N-ACETYLNEURAMINIC ACID, ... | | Authors: | Chen, S.C, Yang, C.S, Ko, T.P, Chen, Y. | | Deposit date: | 2015-04-27 | | Release date: | 2016-06-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Mechanism and inhibition of human UDP-GlcNAc 2-epimerase, the key enzyme in sialic acid biosynthesis.
Sci Rep, 6, 2016
|
|
8GR7
 
 | |
8GR5
 
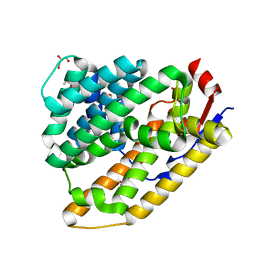 | | Cop4 from Antrodia cinnamomea in apo form | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, AcCop4, GLYCEROL | | Authors: | Chen, S.C, Hsu, C.H. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and Crystal Structures of a Cubebol-Producing Sesquiterpene Synthase from Antrodia cinnamomea .
J.Agric.Food Chem., 71, 2023
|
|
7DS0
 
 | |
7DS1
 
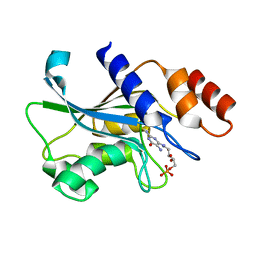 | | Crystal structure of Aspergillus oryzae Rib2 deaminase in complex with DARIPP (C-terminal deletion mutant at pH 6.5) | | Descriptor: | CMP/dCMP-type deaminase domain-containing protein, ZINC ION, [(2~{R},3~{S},4~{S})-5-[[2,5-bis(azanyl)-6-oxidanylidene-1~{H}-pyrimidin-4-yl]amino]-2,3,4-tris(oxidanyl)pentyl] dihydrogen phosphate | | Authors: | Chen, S.C, Liaw, S.H, Hsu, C.H. | | Deposit date: | 2020-12-30 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of Aspergillus oryzae Rib2 deaminase: the functional mechanism involved in riboflavin biosynthesis.
Iucrj, 8, 2021
|
|
7DRZ
 
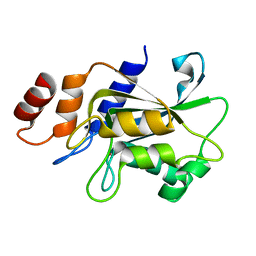 | |
7DRY
 
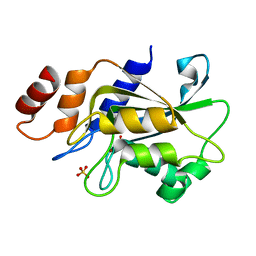 | | Crystal structure of Aspergillus oryzae Rib2 deaminase | | Descriptor: | CMP/dCMP-type deaminase domain-containing protein, SULFATE ION, ZINC ION | | Authors: | Chen, S.C, Liaw, S.H, Hsu, C.H. | | Deposit date: | 2020-12-30 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of Aspergillus oryzae Rib2 deaminase: the functional mechanism involved in riboflavin biosynthesis.
Iucrj, 8, 2021
|
|
3EX8
 
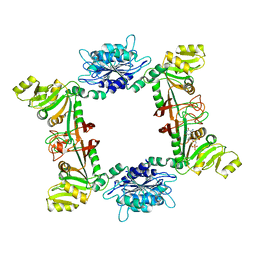 | | Complex structure of bacillus subtilis RibG reduction mechanism in riboflavin biosynthesis | | Descriptor: | Riboflavin biosynthesis protein ribD, ZINC ION, [(2R,3S,4R,5E)-5-[(5-amino-2,6-dioxo-3H-pyrimidin-4-yl)imino]-2,3,4-trihydroxy-pentyl] dihydrogen phosphate | | Authors: | Chen, S.C, Lin, Y.H, Yu, H.C, Liaw, S.H. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Complex structure of Bacillus subtilis RibG: the reduction mechanism during riboflavin biosynthesis.
J.Biol.Chem., 284, 2009
|
|
4NES
 
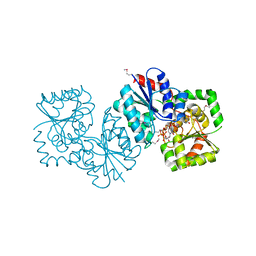 | | Crystal structure of Methanocaldococcus jannaschii UDP-GlcNAc 2-epimerase in complex with UDP-GlcNAc and UDP | | Descriptor: | UDP-N-acetylglucosamine 2-epimerase, URIDINE-5'-DIPHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Chen, S.C, Yang, C.S, Huang, C.H, Chen, Y. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-23 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures of the archaeal UDP-GlcNAc 2-epimerase from Methanocaldococcus jannaschii reveal a conformational change induced by UDP-GlcNAc.
Proteins, 82, 2014
|
|
4NEQ
 
 | | The structure of UDP-GlcNAc 2-epimerase from Methanocaldococcus jannaschii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Chen, S.C, Yang, C.S, Huang, C.H, Chen, Y. | | Deposit date: | 2013-10-29 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of the archaeal UDP-GlcNAc 2-epimerase from Methanocaldococcus jannaschii reveal a conformational change induced by UDP-GlcNAc.
Proteins, 82, 2014
|
|
4Q47
 
 | |
4Q48
 
 | |
6LCJ
 
 | |
6LCL
 
 | |
6LCK
 
 | |
4G3M
 
 | | Complex Structure of Bacillus subtilis RibG: The Deamination Process in Riboflavin Biosynthesis | | Descriptor: | N-(5-amino-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)-5-O-phosphono-beta-D-ribofuranosylamine, Riboflavin biosynthesis protein RibD, ZINC ION, ... | | Authors: | Chen, S.C, Shen, C.Y, Yen, T.M, Yu, H.C, Chang, T.H, Lai, W.L, Liaw, S.H. | | Deposit date: | 2012-07-15 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Evolution of vitamin B(2) biosynthesis: eubacterial RibG and fungal Rib2 deaminases.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5BND
 
 | | Crystal structure of the C-terminal domain of TagH | | Descriptor: | ABC transporter, ATP-binding protein | | Authors: | Chen, S.C, Chen, Y. | | Deposit date: | 2015-05-26 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | SH3-Like Motif-Containing C-terminal Domain of Staphylococcal Teichoic Acid Transporter Suggests Possible Function.
Proteins, 2016
|
|
4QVG
 
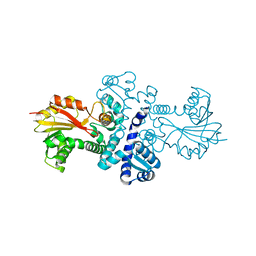 | | Crystal structure of S-adenosylmethionine-dependent methyltransferase SibL in its apo form | | Descriptor: | SibL | | Authors: | Liu, J.S, Chen, S.C, Huang, C.H, Yang, C.S, Chen, Y. | | Deposit date: | 2014-07-15 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of an antibiotics-synthesizing 3-hydroxykynurenine C-methyltransferase
Sci Rep, 5, 2015
|
|
4IWM
 
 | | Crystal Structure of the Conserved Hypothetical Protein MJ0927 from Methanocaldococcus jannaschii (in P21 form) | | Descriptor: | UPF0135 protein MJ0927 | | Authors: | Kuan, S.M, Chen, S.C, Yang, C.S, Chen, Y.R, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-01-24 | | Release date: | 2014-01-29 | | Last modified: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a conserved hypothetical protein MJ0927 from Methanocaldococcus jannaschii reveals a novel quaternary assembly in the Nif3 family.
Biomed Res Int, 2014, 2014
|
|
4IWG
 
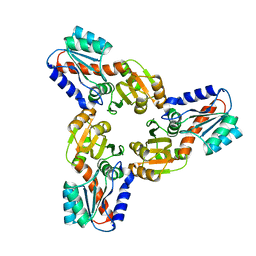 | | Crystal Structure of the Conserved Hypothetical Protein MJ0927 from Methanocaldococcus jannaschii (in C2221 form) | | Descriptor: | UPF0135 protein MJ0927 | | Authors: | Kuan, S.M, Chen, S.C, Yang, C.S, Chen, Y.R, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-01-23 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Crystal structure of a conserved hypothetical protein MJ0927 from Methanocaldococcus jannaschii reveals a novel quaternary assembly in the Nif3 family.
Biomed Res Int, 2014, 2014
|
|
5XUX
 
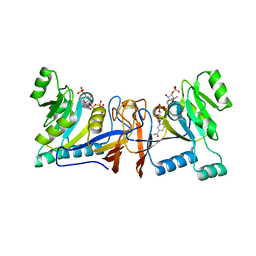 | | Crystal structure of Rib7 from Methanosarcina mazei | | Descriptor: | Conserved protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5XV5
 
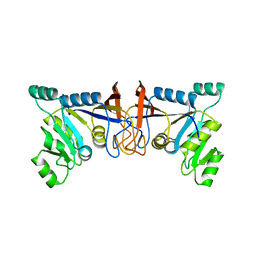 | | Crystal structure of Rib7 mutant S88E from Methanosarcina mazei | | Descriptor: | Conserved protein | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5XV0
 
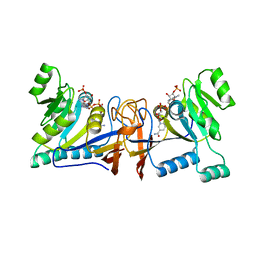 | | Crystal structure of Rib7 mutant D33N from Methanosarcina mazei | | Descriptor: | Conserved protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5XV2
 
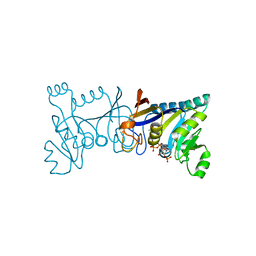 | | Crystal structure of Rib7 mutant D33A from Methanosarcina mazei | | Descriptor: | Conserved protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
4X3Q
 
 | | Crystal structure of S-adenosylmethionine-dependent methyltransferase SibL in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SibL | | Authors: | liu, J.S, Chen, S.C, Yang, C.S, Huang, C.H, Chen, Y. | | Deposit date: | 2014-12-01 | | Release date: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Crystal structure of S-adenosylmethionine-dependent methyltransferase SibL in complex with SAH
To Be Published, 2015
|
|
