6RZ3
 
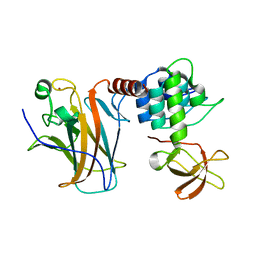 | | Crystal structure of a complex between the DNA-binding domain of p53 and the carboxyl-terminal conserved region of iASPP | | Descriptor: | Cellular tumor antigen p53, RelA-associated inhibitor, ZINC ION | | Authors: | Chen, S, Ren, J, Jones, E.Y, Lu, X. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.23 Å) | | Cite: | iASPP mediates p53 selectivity through a modular mechanism fine-tuning DNA recognition.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1O3S
 
 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Chen, S, Ebright, R.H, Berman, H.M. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: Alteration of DNA Binding Specificity Through Alteration of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
1O3Q
 
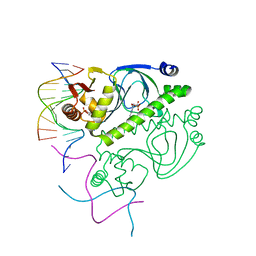 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Chen, S, Vojtechovsky, J, Parkinson, G.N, Ebright, R.H, Berman, H.M. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: DNA Binding Specificity Based on Energetics of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
1O3R
 
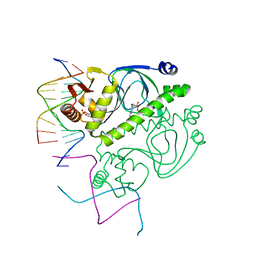 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Chen, S, Vojtechovsky, J, Parkinson, G.N, Ebright, R.H, Berman, H.M. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: DNA Binding Specificity Based on Energetics of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
1O3T
 
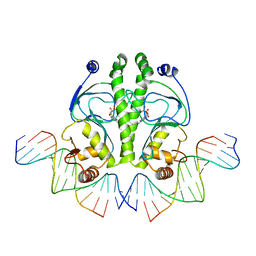 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | Descriptor: | 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Chen, S, Vojtechovsky, J, Parkinson, G.N, Ebright, R.H, Berman, H.M. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex:
DNA Binding Specificity Based on Energetics of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
7N7J
 
 | | Crystal Structure of PI5P4KIIAlpha complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, ... | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N6Z
 
 | | Crystal Structure of PI5P4KIIAlpha | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7K
 
 | | Crystal Structure of PI5P4KIIAlpha complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, ... | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N71
 
 | | Crystal Structure of PI5P4KIIAlpha | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7O
 
 | | Crystal Structure of PI5P4KIIAlpha complex with Palbociclib | | Descriptor: | 6-ACETYL-8-CYCLOPENTYL-5-METHYL-2-[(5-PIPERAZIN-1-YLPYRIDIN-2-YL)AMINO]PYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7N
 
 | | Crystal Structure of PI5P4KIIAlpha complex with Volasertib | | Descriptor: | N-{trans-4-[4-(cyclopropylmethyl)piperazin-1-yl]cyclohexyl}-4-{[(7R)-7-ethyl-5-methyl-8-(1-methylethyl)-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxybenzamide, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N81
 
 | | Crystal Structure of PI5P4KIIBeta complex with CC260 | | Descriptor: | (7R)-8-cyclopentyl-7-(cyclopentylmethyl)-2-[(3,5-dichloro-4-hydroxyphenyl)amino]-5-methyl-7,8-dihydropteridin-6(5H)-one, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-11 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N80
 
 | | Crystal Structure of PI5P4KIIBeta | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-11 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7L
 
 | | Crystal Structure of PI5P4KIIAlpha complex with BI-D1870 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7-dimethyl-8-(3-methylbutyl)-7,8-dihydropteridin-6(5H)-one, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7M
 
 | | Crystal Structure of PI5P4KIIAlpha complex with BI-2536 | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7P8W
 
 | | Human erythrocyte catalase cryoEM | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chen, S, Li, J, Vinothkumar, K.R, Henderson, R. | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Interaction of human erythrocyte catalase with air-water interface in cryoEM.
Microscopy (Oxf), 71, 2022
|
|
2PWX
 
 | | Crystal structure of G11A mutant of SARS-CoV 3C-like protease | | Descriptor: | 3C-like proteinase | | Authors: | Chen, S, Hu, T, Jiang, H, Shen, X. | | Deposit date: | 2007-05-14 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutation of Gly-11 on the dimer interface results in the complete crystallographic dimer dissociation of severe acute respiratory syndrome coronavirus 3C-like protease: crystal structure with molecular dynamics simulations.
J.Biol.Chem., 283, 2008
|
|
6NQ3
 
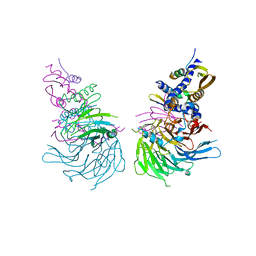 | | Crystal Structure of a SUZ12-RBBP4-PHF19-JARID2 Heterotetrameric Complex | | Descriptor: | Histone-binding protein RBBP4, PHD finger protein 19, Polycomb protein SUZ12, ... | | Authors: | Chen, S, Jiao, L, Liu, X. | | Deposit date: | 2019-01-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | A Dimeric Structural Scaffold for PRC2-PCL Targeting to CpG Island Chromatin.
Mol.Cell, 77, 2020
|
|
1N0F
 
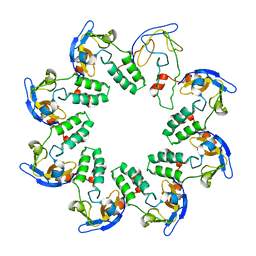 | | CRYSTAL STRUCTURE OF A CELL DIVISION AND CELL WALL BIOSYNTHESIS PROTEIN UPF0040 FROM MYCOPLASMA PNEUMONIAE: INDICATION OF A NOVEL FOLD WITH A POSSIBLE NEW CONSERVED SEQUENCE MOTIF | | Descriptor: | Protein mraZ | | Authors: | Chen, S, Jancrick, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-10-13 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a protein associated with cell division from Mycoplasma pneumoniae (GI: 13508053): a novel fold with a conserved sequence motif.
Proteins, 55, 2004
|
|
1N0E
 
 | | CRYSTAL STRUCTURE OF A CELL DIVISION AND CELL WALL BIOSYNTHESIS PROTEIN UPF0040 FROM MYCOPLASMA PNEUMONIAE: INDICATION OF A NOVEL FOLD WITH A POSSIBLE NEW CONSERVED SEQUENCE MOTIF | | Descriptor: | Protein mraZ | | Authors: | Chen, S, Jancrick, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-10-13 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a protein associated with cell division from Mycoplasma pneumoniae (GI: 13508053): a novel fold with a conserved sequence motif.
Proteins, 55, 2004
|
|
1N0G
 
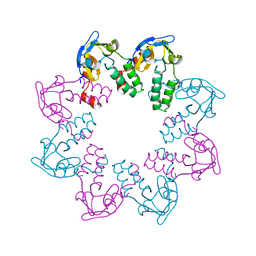 | | Crystal Structure of A Cell Division and Cell Wall Biosynthesis Protein UPF0040 from Mycoplasma pneumoniae: Indication of A Novel Fold with A Possible New Conserved Sequence Motif | | Descriptor: | Protein mraZ | | Authors: | Chen, S, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-10-13 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a protein associated with cell division from Mycoplasma pneumoniae (GI: 13508053): a novel fold with a conserved sequence motif.
Proteins, 55, 2004
|
|
4JBW
 
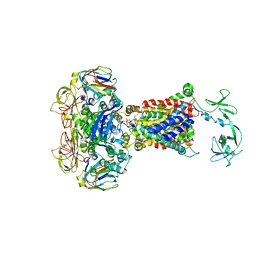 | | Crystal structure of E. coli maltose transporter MalFGK2 in complex with its regulatory protein EIIAglc | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Glucose-specific phosphotransferase enzyme IIA component, Maltose transport system permease protein MalF, ... | | Authors: | Chen, S, Oldham, M.L, Davidson, A.L, Chen, J. | | Deposit date: | 2013-02-20 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.913 Å) | | Cite: | Carbon catabolite repression of the maltose transporter revealed by X-ray crystallography.
Nature, 499, 2013
|
|
2AHD
 
 | |
5DAH
 
 | |
5DAG
 
 | |
