4V0X
 
 | | The crystal structure of mouse PP1G in complex with truncated human PPP1R15B (631-684) | | Descriptor: | MANGANESE (II) ION, PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 15B, PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
4V0W
 
 | | The crystal structure of mouse PP1G in complex with truncated human PPP1R15B (631-669) | | Descriptor: | MANGANESE (II) ION, PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 15B, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
4V0U
 
 | | The crystal structure of ternary PP1G-PPP1R15B and G-actin complex | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (7.88 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
4V0V
 
 | | The crystal structure of mouse PP1G in complex with truncated human PPP1R15B (631-660) | | Descriptor: | MANGANESE (II) ION, PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 15B, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT, ... | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
7E2Q
 
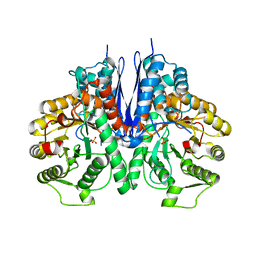 | | Crystal structure of Mycoplasma pneumoniae Enolase | | Descriptor: | Enolase, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Xiong, Q, Shao, G, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
7E2P
 
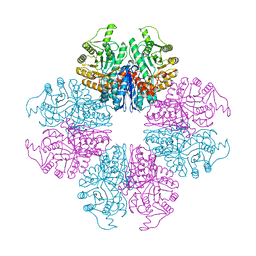 | | The Crystal Structure of Mycoplasma bovis enolase | | Descriptor: | Enolase | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Shao, G, Xiong, Q, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
5WY4
 
 | |
4PBP
 
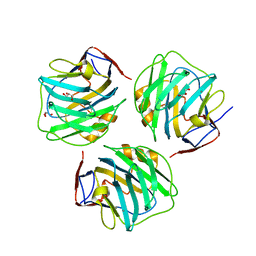 | | crystal structure of zebrafish short-chain pentraxin protein | | Descriptor: | C-reactive protein, CALCIUM ION, GLYCEROL | | Authors: | Chen, R, Qi, J.X, George, F.G, Xia, C. | | Deposit date: | 2014-04-13 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Crystal structures for short-chain pentraxin from zebrafish demonstrate a cyclic trimer with new recognition and effector faces.
J.Struct.Biol., 189, 2015
|
|
4PBO
 
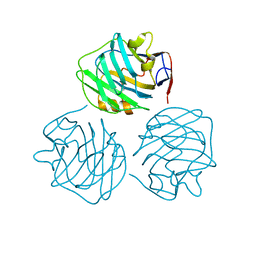 | |
2GH8
 
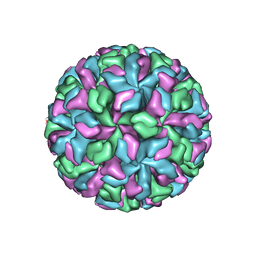 | | X-ray structure of a native calicivirus | | Descriptor: | Capsid protein | | Authors: | Chen, R. | | Deposit date: | 2006-03-26 | | Release date: | 2006-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a native calicivirus: Structural insights into antigenic diversity and host specificity.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
8IK1
 
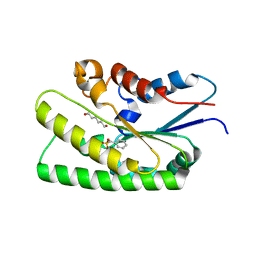 | | EstL5 in complex of PMSF | | Descriptor: | GDSL family lipase, HEXANE-1,6-DIOL | | Authors: | Chen, R, Zhang, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2023-03-29 | | Last modified: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | EstL5 in complex of PMSF
To Be Published
|
|
6J36
 
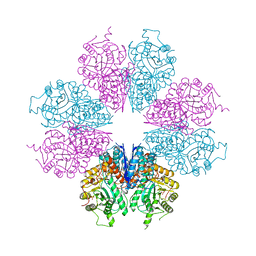 | | crystal structure of Mycoplasma hyopneumoniae Enolase | | Descriptor: | Enolase, GLYCEROL, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Xie, X, Feng, Z, Wang, W, Ran, T, Zhang, W, Xiang, Q, Shao, G. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Featured Species-Specific Loops Are Found in the Crystal Structure ofMhpEno, a Cell Surface Adhesin FromMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 9, 2019
|
|
5XPV
 
 | | Structure of the V domain of amphioxus IgVJ-C2 | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
5XPW
 
 | | Structure of amphioxus IgVJ-C2 molecule | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
5WYL
 
 | |
5WY3
 
 | |
5WYG
 
 | | The crystal structure of the apo form of Mtb MazF | | Descriptor: | Probable endoribonuclease MazF7 | | Authors: | Xie, W, Chen, R, Tu, J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Structure of the MazF-mt9 toxin, a tRNA-specific endonuclease from Mycobacterium tuberculosis
Biochem. Biophys. Res. Commun., 486, 2017
|
|
8YLE
 
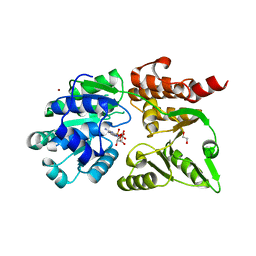 | | Crystal structure of Werner syndrome helicase complexed with AMP-PCP | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Yang, Y, Fu, L, Sun, X, Cheng, H, Chen, R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of werner syndrome helicase complexed with AMP-PCP at 1.86 Angstroms resolution.
To Be Published
|
|
4YVW
 
 | | crystal structure of an enterovirus 71/coxsackievirus A16 chimeric virus-like particle | | Descriptor: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3 | | Authors: | Chen, R, Lyu, K. | | Deposit date: | 2015-03-20 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal Structures of Yeast-Produced Enterovirus 71 and Enterovirus 71/Coxsackievirus A16 Chimeric Virus-Like Particles Provide the Structural Basis for Novel Vaccine Design against Hand-Foot-and-Mouth Disease
J.Virol., 89, 2015
|
|
4YVS
 
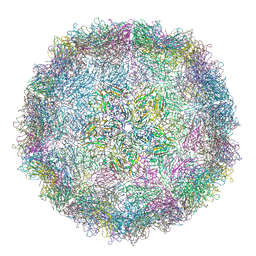 | | crystal structure of the virus-like particle of a c4 strain EV71 | | Descriptor: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3 | | Authors: | Chen, R, Lyu, K. | | Deposit date: | 2015-03-20 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Crystal Structures of Yeast-Produced Enterovirus 71 and Enterovirus 71/Coxsackievirus A16 Chimeric Virus-Like Particles Provide the Structural Basis for Novel Vaccine Design against Hand-Foot-and-Mouth Disease
J.Virol., 89, 2015
|
|
4RQP
 
 | |
4RS5
 
 | |
4RR3
 
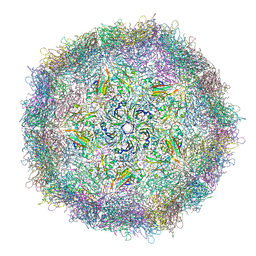 | | Crystal structure of a recombinant EV71 virus particle | | Descriptor: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3 | | Authors: | Chen, R, Lyu, K. | | Deposit date: | 2014-11-06 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Crystal structures of enterovirus 71 (EV71) recombinant virus particles provide insights into vaccine design.
J.Biol.Chem., 290, 2015
|
|
7EZG
 
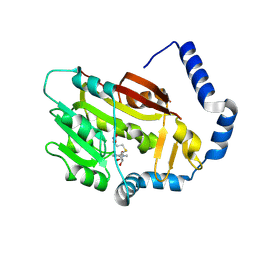 | | The structure of the human METTL6 enzyme in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Xie, W, Chen, R, Zhou, J, Liu, L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human METTL6, the m 3 C methyltransferase.
Commun Biol, 4, 2021
|
|
6L29
 
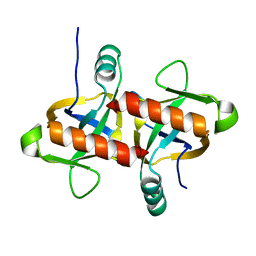 | | The structure of the MazF-mt1 mutant | | Descriptor: | mRNA interferase | | Authors: | Xie, W, Chen, R, Zhou, J. | | Deposit date: | 2019-10-02 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3000052 Å) | | Cite: | Conserved Conformational Changes in the Regulation ofMycobacterium tuberculosisMazEF-mt1.
Acs Infect Dis., 6, 2020
|
|
